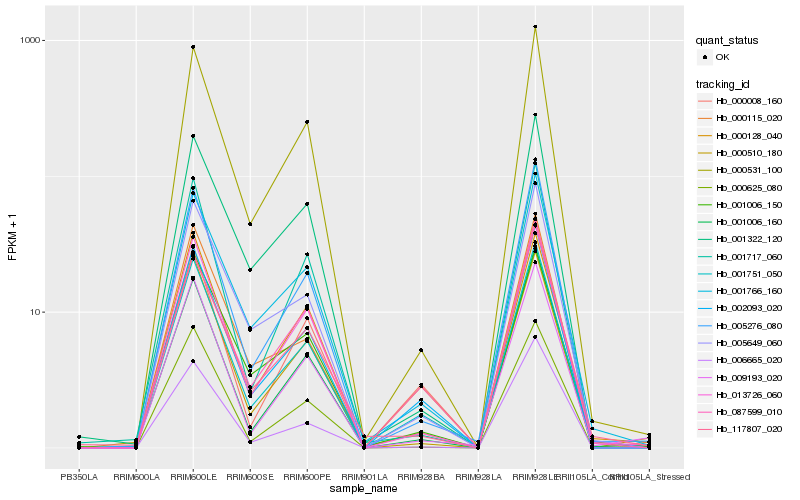
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001751_050 |
0.0 |
- |
- |
PREDICTED: phytol kinase 1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_002093_020 |
0.0218710635 |
- |
- |
hypothetical protein POPTR_0008s02960g [Populus trichocarpa] |
| 3 |
Hb_000531_100 |
0.0661914424 |
- |
- |
Photosystem I reaction center subunit III, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_013726_060 |
0.0665748886 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2 [Jatropha curcas] |
| 5 |
Hb_005276_080 |
0.0725096099 |
- |
- |
PREDICTED: chloroplast stem-loop binding protein of 41 kDa a, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001766_160 |
0.072606424 |
- |
- |
Serine/threonine-protein kinase SAPK1 [Gossypium arboreum] |
| 7 |
Hb_000008_160 |
0.0730962203 |
- |
- |
carboxy-lyase, putative [Ricinus communis] |
| 8 |
Hb_005649_060 |
0.0733553545 |
- |
- |
PREDICTED: uncharacterized protein LOC105644161 [Jatropha curcas] |
| 9 |
Hb_000115_020 |
0.0832807421 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger family protein [Populus trichocarpa] |
| 10 |
Hb_009193_020 |
0.0834959841 |
- |
- |
PREDICTED: epoxide hydrolase 3 [Jatropha curcas] |
| 11 |
Hb_001717_060 |
0.0847559981 |
- |
- |
PREDICTED: xylogen-like protein 11 [Gossypium raimondii] |
| 12 |
Hb_000625_080 |
0.0887418115 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, leaf isozyme, chloroplastic [Jatropha curcas] |
| 13 |
Hb_006665_020 |
0.0887819023 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 14 |
Hb_087599_010 |
0.0896693063 |
- |
- |
PREDICTED: tropinone reductase homolog [Solanum tuberosum] |
| 15 |
Hb_117807_020 |
0.0897666295 |
- |
- |
methyltransferase, putative [Ricinus communis] |
| 16 |
Hb_000510_180 |
0.089960623 |
- |
- |
unknown [Lotus japonicus] |
| 17 |
Hb_001006_160 |
0.0900523773 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 18 |
Hb_000128_040 |
0.0904441757 |
- |
- |
PREDICTED: uncharacterized protein LOC105641228 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001322_120 |
0.0908386619 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 20 |
Hb_001006_150 |
0.0919281328 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |