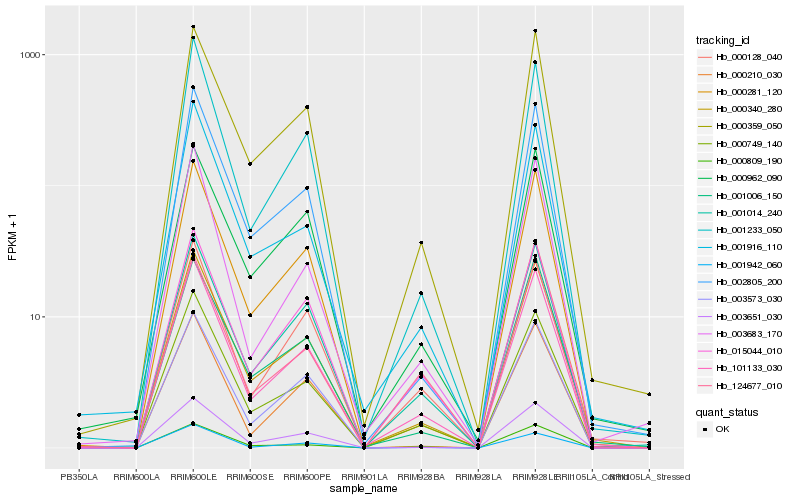
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_101133_030 |
0.0 |
- |
- |
DUF26 domain-containing protein 1 precursor, putative [Ricinus communis] |
| 2 |
Hb_000340_280 |
0.0518990447 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000749_140 |
0.0526498227 |
- |
- |
PREDICTED: 2-succinylbenzoate--CoA ligase, chloroplastic/peroxisomal [Jatropha curcas] |
| 4 |
Hb_001014_240 |
0.0632180512 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.1 [Jatropha curcas] |
| 5 |
Hb_000359_050 |
0.0642755964 |
- |
- |
Oxygen-evolving enhancer protein 2, chloroplast precursor, putative [Ricinus communis] |
| 6 |
Hb_001233_050 |
0.0649075838 |
- |
- |
Magnesium-protoporphyrin IX monomethyl ester [oxidative] cyclase, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_003651_030 |
0.0661126923 |
- |
- |
lyase, putative [Ricinus communis] |
| 8 |
Hb_000281_120 |
0.0716755543 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 9 |
Hb_002805_200 |
0.0728440855 |
- |
- |
PREDICTED: serine--glyoxylate aminotransferase [Nelumbo nucifera] |
| 10 |
Hb_015044_010 |
0.0733331381 |
- |
- |
hypothetical protein JCGZ_25130 [Jatropha curcas] |
| 11 |
Hb_001942_060 |
0.0746704303 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003683_170 |
0.076918983 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000128_040 |
0.0774464238 |
- |
- |
PREDICTED: uncharacterized protein LOC105641228 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001916_110 |
0.0785859939 |
- |
- |
PREDICTED: uncharacterized protein LOC105641510 [Jatropha curcas] |
| 15 |
Hb_000809_190 |
0.0813123792 |
- |
- |
PREDICTED: myosin-6-like [Jatropha curcas] |
| 16 |
Hb_124677_010 |
0.0827005346 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001006_150 |
0.0839827209 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 18 |
Hb_000210_030 |
0.0865947646 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP6 [Jatropha curcas] |
| 19 |
Hb_000962_090 |
0.0920985631 |
- |
- |
omega-6 fatty acid desaturase [Hevea brasiliensis] |
| 20 |
Hb_003573_030 |
0.0923289687 |
- |
- |
PREDICTED: alkane hydroxylase MAH1-like [Jatropha curcas] |