| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
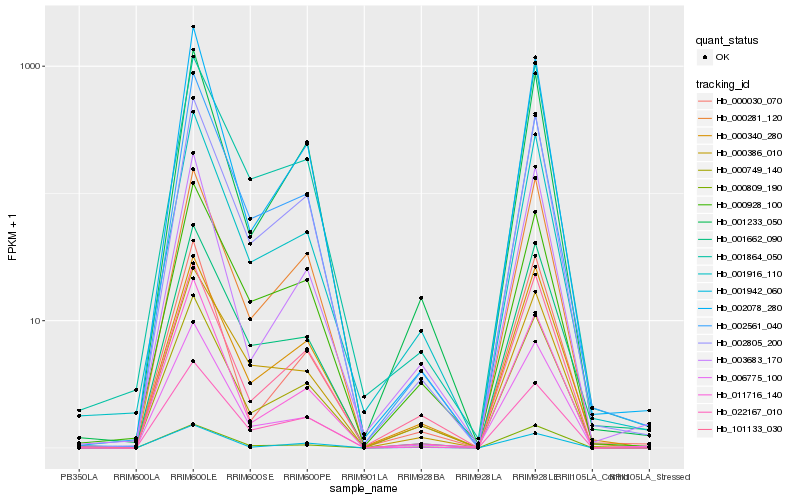
Hb_001916_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641510 [Jatropha curcas] |
| 2 |
Hb_000749_140 |
0.0549735741 |
- |
- |
PREDICTED: 2-succinylbenzoate--CoA ligase, chloroplastic/peroxisomal [Jatropha curcas] |
| 3 |
Hb_006775_100 |
0.063994974 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 4 |
Hb_002805_200 |
0.0723551947 |
- |
- |
PREDICTED: serine--glyoxylate aminotransferase [Nelumbo nucifera] |
| 5 |
Hb_000340_280 |
0.0774297334 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000928_100 |
0.077971694 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 7 |
Hb_101133_030 |
0.0785859939 |
- |
- |
DUF26 domain-containing protein 1 precursor, putative [Ricinus communis] |
| 8 |
Hb_001864_050 |
0.0844645468 |
- |
- |
chloroplast oxygen-evolving enhancer protein [Manihot esculenta] |
| 9 |
Hb_003683_170 |
0.0847724892 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001233_050 |
0.0856129232 |
- |
- |
Magnesium-protoporphyrin IX monomethyl ester [oxidative] cyclase, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_001662_090 |
0.0860013841 |
- |
- |
PREDICTED: thiosulfate sulfurtransferase 16, chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_000809_190 |
0.0887737415 |
- |
- |
PREDICTED: myosin-6-like [Jatropha curcas] |
| 13 |
Hb_002561_040 |
0.0908202717 |
- |
- |
Photosynthetic electron transfer C [Theobroma cacao] |
| 14 |
Hb_000386_010 |
0.0915689091 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002078_280 |
0.0924874435 |
- |
- |
chlorophyll a/b-binding protein type II precursor [Populus trichocarpa] |
| 16 |
Hb_000030_070 |
0.0939466783 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_022167_010 |
0.0953116414 |
- |
- |
Kinase, putative isoform 2 [Theobroma cacao] |
| 18 |
Hb_001942_060 |
0.0960262231 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000281_120 |
0.1011424658 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 20 |
Hb_011716_140 |
0.101599854 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein J [Jatropha curcas] |