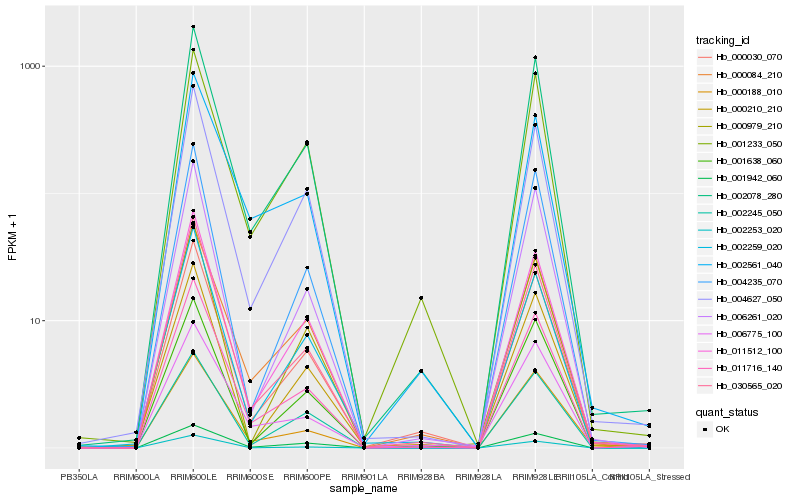
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011716_140 |
0.0 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein J [Jatropha curcas] |
| 2 |
Hb_002078_280 |
0.0359772804 |
- |
- |
chlorophyll a/b-binding protein type II precursor [Populus trichocarpa] |
| 3 |
Hb_030565_020 |
0.0568360962 |
- |
- |
PREDICTED: protein ECERIFERUM 26 [Jatropha curcas] |
| 4 |
Hb_002253_020 |
0.0585479 |
- |
- |
PREDICTED: callose synthase 5 [Jatropha curcas] |
| 5 |
Hb_011512_100 |
0.0590898482 |
- |
- |
PREDICTED: uncharacterized protein LOC105650382 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004627_050 |
0.0653459824 |
- |
- |
Chlorophyll a-b binding protein 6A [Morus notabilis] |
| 7 |
Hb_002259_020 |
0.0767540613 |
- |
- |
PREDICTED: uncharacterized protein LOC105646333 [Jatropha curcas] |
| 8 |
Hb_002561_040 |
0.0777896679 |
- |
- |
Photosynthetic electron transfer C [Theobroma cacao] |
| 9 |
Hb_006261_020 |
0.079277425 |
- |
- |
Vacuolar cation/proton exchanger 1a, putative [Ricinus communis] |
| 10 |
Hb_004235_070 |
0.0813499785 |
- |
- |
acyl carrier protein, putative [Ricinus communis] |
| 11 |
Hb_006775_100 |
0.0819874705 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 12 |
Hb_000188_010 |
0.0826751586 |
- |
- |
PREDICTED: cytochrome P450 94C1-like [Jatropha curcas] |
| 13 |
Hb_001638_060 |
0.0844255931 |
- |
- |
PREDICTED: rho GTPase-activating protein 3-like isoform X1 [Populus euphratica] |
| 14 |
Hb_001233_050 |
0.0934935778 |
- |
- |
Magnesium-protoporphyrin IX monomethyl ester [oxidative] cyclase, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_000210_210 |
0.0938318241 |
- |
- |
PREDICTED: uncharacterized protein LOC105635822 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000030_070 |
0.095524193 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002245_050 |
0.0965883164 |
- |
- |
Oxygen-evolving enhancer protein 3-1, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_000979_210 |
0.0967565163 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001942_060 |
0.0969446287 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000084_210 |
0.0974482379 |
- |
- |
hypothetical protein POPTR_0016s08050g [Populus trichocarpa] |