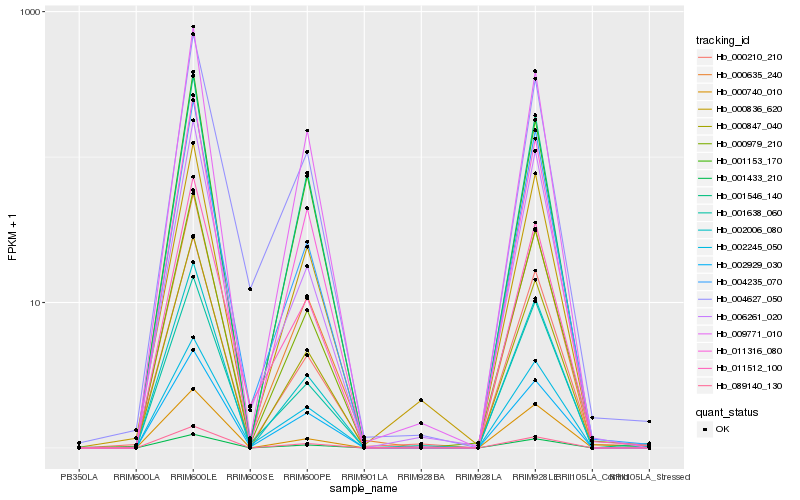
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000979_210 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002006_080 |
0.0428618108 |
- |
- |
PREDICTED: CRAL-TRIO domain-containing protein YKL091C-like isoform X2 [Citrus sinensis] |
| 3 |
Hb_000847_040 |
0.0429163979 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 9 [Jatropha curcas] |
| 4 |
Hb_011316_080 |
0.0462626605 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 5 |
Hb_000210_210 |
0.050830265 |
- |
- |
PREDICTED: uncharacterized protein LOC105635822 isoform X1 [Jatropha curcas] |
| 6 |
Hb_089140_130 |
0.0571298373 |
- |
- |
hypothetical protein POPTR_0006s14510g [Populus trichocarpa] |
| 7 |
Hb_000635_240 |
0.0579378034 |
- |
- |
chlorophyll a/b-binding protein type II precursor [Populus trichocarpa] |
| 8 |
Hb_004235_070 |
0.0587436381 |
- |
- |
acyl carrier protein, putative [Ricinus communis] |
| 9 |
Hb_011512_100 |
0.0593334153 |
- |
- |
PREDICTED: uncharacterized protein LOC105650382 isoform X1 [Jatropha curcas] |
| 10 |
Hb_009771_010 |
0.0622383711 |
- |
- |
PREDICTED: glycerol-3-phosphate 2-O-acyltransferase 6 [Populus euphratica] |
| 11 |
Hb_001153_170 |
0.0638010153 |
- |
- |
PREDICTED: protein HOTHEAD [Populus euphratica] |
| 12 |
Hb_006261_020 |
0.0660792487 |
- |
- |
Vacuolar cation/proton exchanger 1a, putative [Ricinus communis] |
| 13 |
Hb_001546_140 |
0.0674938142 |
- |
- |
Auxin-binding protein ABP19a precursor, putative [Ricinus communis] |
| 14 |
Hb_004627_050 |
0.0689825747 |
- |
- |
Chlorophyll a-b binding protein 6A [Morus notabilis] |
| 15 |
Hb_001433_210 |
0.0696272055 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 20 [Jatropha curcas] |
| 16 |
Hb_000740_010 |
0.0698176075 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 17 |
Hb_001638_060 |
0.0701701016 |
- |
- |
PREDICTED: rho GTPase-activating protein 3-like isoform X1 [Populus euphratica] |
| 18 |
Hb_002929_030 |
0.0710262355 |
- |
- |
Serine/threonine-protein kinase-transforming protein raf, putative [Ricinus communis] |
| 19 |
Hb_002245_050 |
0.0731065507 |
- |
- |
Oxygen-evolving enhancer protein 3-1, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_000836_620 |
0.0744551441 |
- |
- |
PREDICTED: protochlorophyllide reductase, chloroplastic [Jatropha curcas] |