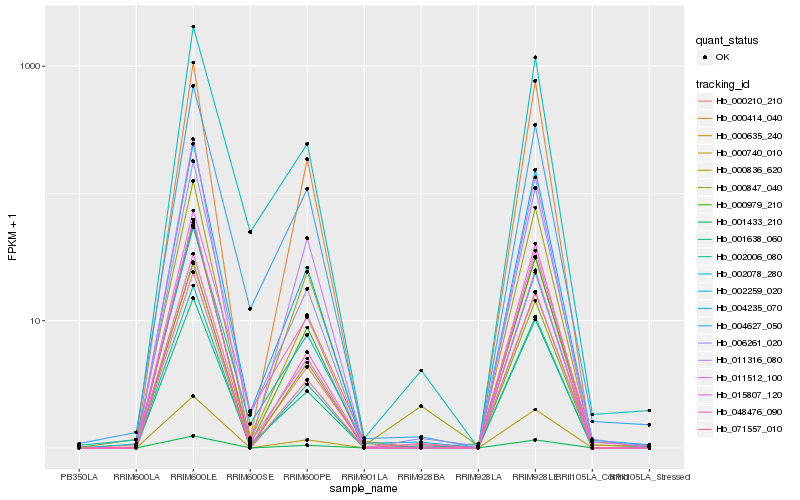
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000210_210 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635822 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000979_210 |
0.050830265 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001638_060 |
0.0532895041 |
- |
- |
PREDICTED: rho GTPase-activating protein 3-like isoform X1 [Populus euphratica] |
| 4 |
Hb_004235_070 |
0.0534189408 |
- |
- |
acyl carrier protein, putative [Ricinus communis] |
| 5 |
Hb_006261_020 |
0.0534473467 |
- |
- |
Vacuolar cation/proton exchanger 1a, putative [Ricinus communis] |
| 6 |
Hb_002006_080 |
0.0557814018 |
- |
- |
PREDICTED: CRAL-TRIO domain-containing protein YKL091C-like isoform X2 [Citrus sinensis] |
| 7 |
Hb_000836_620 |
0.0623961493 |
- |
- |
PREDICTED: protochlorophyllide reductase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000740_010 |
0.0646537181 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 9 |
Hb_000847_040 |
0.0662232883 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 9 [Jatropha curcas] |
| 10 |
Hb_000635_240 |
0.0691972841 |
- |
- |
chlorophyll a/b-binding protein type II precursor [Populus trichocarpa] |
| 11 |
Hb_071557_010 |
0.0709469973 |
- |
- |
hypothetical protein POPTR_0008s06270g [Populus trichocarpa] |
| 12 |
Hb_011316_080 |
0.0712993779 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 13 |
Hb_015807_120 |
0.0721434098 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 3, chloroplastic [Jatropha curcas] |
| 14 |
Hb_011512_100 |
0.0727359711 |
- |
- |
PREDICTED: uncharacterized protein LOC105650382 isoform X1 [Jatropha curcas] |
| 15 |
Hb_048476_090 |
0.0739387003 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000414_040 |
0.0749671391 |
- |
- |
hypothetical protein POPTR_0009s13250g [Populus trichocarpa] |
| 17 |
Hb_004627_050 |
0.0762248056 |
- |
- |
Chlorophyll a-b binding protein 6A [Morus notabilis] |
| 18 |
Hb_002078_280 |
0.0775518887 |
- |
- |
chlorophyll a/b-binding protein type II precursor [Populus trichocarpa] |
| 19 |
Hb_002259_020 |
0.0780242704 |
- |
- |
PREDICTED: uncharacterized protein LOC105646333 [Jatropha curcas] |
| 20 |
Hb_001433_210 |
0.0809500268 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 20 [Jatropha curcas] |