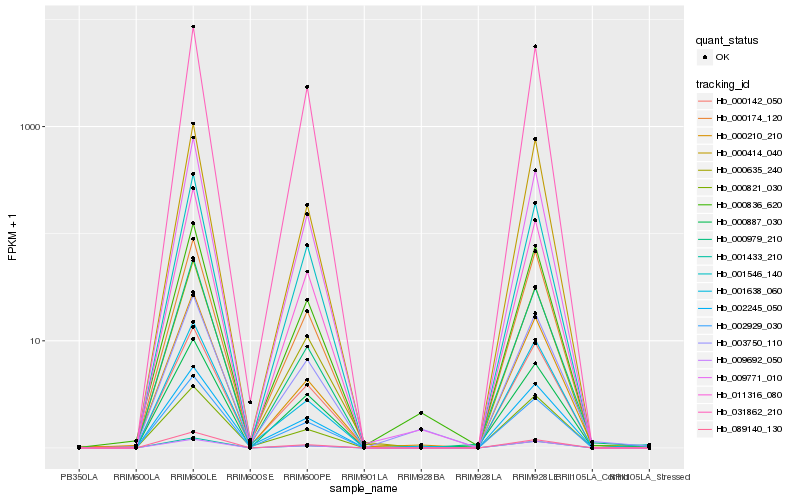
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000836_620 |
0.0 |
- |
- |
PREDICTED: protochlorophyllide reductase, chloroplastic [Jatropha curcas] |
| 2 |
Hb_003750_110 |
0.0541854165 |
- |
- |
PREDICTED: aldose 1-epimerase-like [Jatropha curcas] |
| 3 |
Hb_002929_030 |
0.0558644845 |
- |
- |
Serine/threonine-protein kinase-transforming protein raf, putative [Ricinus communis] |
| 4 |
Hb_001433_210 |
0.0613510536 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 20 [Jatropha curcas] |
| 5 |
Hb_000635_240 |
0.0621856638 |
- |
- |
chlorophyll a/b-binding protein type II precursor [Populus trichocarpa] |
| 6 |
Hb_000210_210 |
0.0623961493 |
- |
- |
PREDICTED: uncharacterized protein LOC105635822 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000142_050 |
0.0666177471 |
- |
- |
PREDICTED: beta-glucosidase 18-like isoform X1 [Populus euphratica] |
| 8 |
Hb_009771_010 |
0.0669331787 |
- |
- |
PREDICTED: glycerol-3-phosphate 2-O-acyltransferase 6 [Populus euphratica] |
| 9 |
Hb_000414_040 |
0.0678283046 |
- |
- |
hypothetical protein POPTR_0009s13250g [Populus trichocarpa] |
| 10 |
Hb_001546_140 |
0.0685158732 |
- |
- |
Auxin-binding protein ABP19a precursor, putative [Ricinus communis] |
| 11 |
Hb_000174_120 |
0.068894765 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 12 |
Hb_002245_050 |
0.0694787075 |
- |
- |
Oxygen-evolving enhancer protein 3-1, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_001638_060 |
0.0699610306 |
- |
- |
PREDICTED: rho GTPase-activating protein 3-like isoform X1 [Populus euphratica] |
| 14 |
Hb_000887_030 |
0.0706656993 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 15 |
Hb_011316_080 |
0.071194695 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 16 |
Hb_000821_030 |
0.0713691079 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN-2 [Jatropha curcas] |
| 17 |
Hb_009692_050 |
0.0728879773 |
- |
- |
- |
| 18 |
Hb_000979_210 |
0.0744551441 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_031862_210 |
0.0778486917 |
- |
- |
hypothetical protein JCGZ_24955 [Jatropha curcas] |
| 20 |
Hb_089140_130 |
0.0779273859 |
- |
- |
hypothetical protein POPTR_0006s14510g [Populus trichocarpa] |