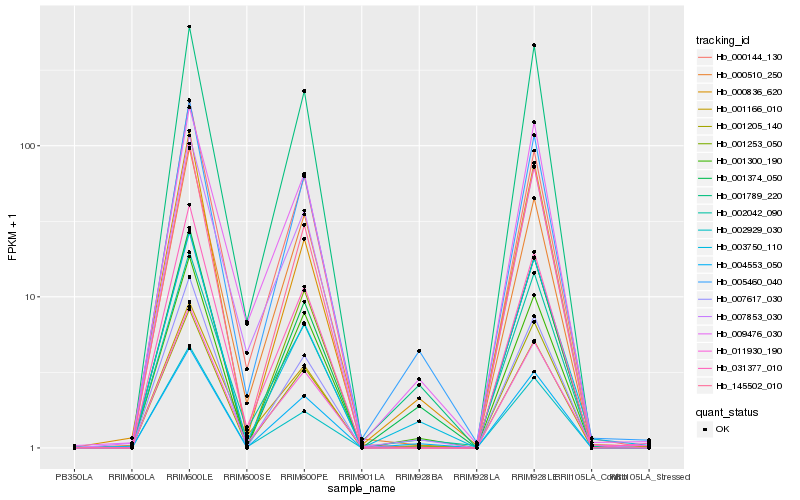
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005460_040 |
0.0 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 2 [Jatropha curcas] |
| 2 |
Hb_004553_050 |
0.0369545251 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g10020 [Jatropha curcas] |
| 3 |
Hb_001374_050 |
0.0530182411 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.3 [Jatropha curcas] |
| 4 |
Hb_002042_090 |
0.0657475534 |
- |
- |
PREDICTED: root phototropism protein 3-like [Jatropha curcas] |
| 5 |
Hb_001300_190 |
0.0691099498 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001789_220 |
0.0729946538 |
- |
- |
PREDICTED: BAHD acyltransferase DCR-like [Jatropha curcas] |
| 7 |
Hb_001166_010 |
0.0738172107 |
- |
- |
S-locus-specific glycoprotein S13 precursor, putative [Ricinus communis] |
| 8 |
Hb_003750_110 |
0.074803178 |
- |
- |
PREDICTED: aldose 1-epimerase-like [Jatropha curcas] |
| 9 |
Hb_011930_190 |
0.0771806527 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 17 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001253_050 |
0.0801357276 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 11 |
Hb_001205_140 |
0.080283196 |
- |
- |
PREDICTED: cytochrome P450 90B1 [Jatropha curcas] |
| 12 |
Hb_007853_030 |
0.0811901383 |
- |
- |
structural molecule, putative [Ricinus communis] |
| 13 |
Hb_009476_030 |
0.0818273603 |
- |
- |
fatty acid desaturase [Manihot esculenta] |
| 14 |
Hb_002929_030 |
0.0824058514 |
- |
- |
Serine/threonine-protein kinase-transforming protein raf, putative [Ricinus communis] |
| 15 |
Hb_031377_010 |
0.0837619863 |
transcription factor |
TF Family: C2C2-YABBY |
unknown [Populus trichocarpa] |
| 16 |
Hb_000144_130 |
0.0841448731 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000836_620 |
0.0863811733 |
- |
- |
PREDICTED: protochlorophyllide reductase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_145502_010 |
0.0868086036 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB82-like [Jatropha curcas] |
| 19 |
Hb_000510_250 |
0.0869971269 |
transcription factor |
TF Family: AUX/IAA |
auxin-induced protein aux28 [Populus trichocarpa] |
| 20 |
Hb_007617_030 |
0.0871966981 |
- |
- |
hypothetical protein JCGZ_26801 [Jatropha curcas] |