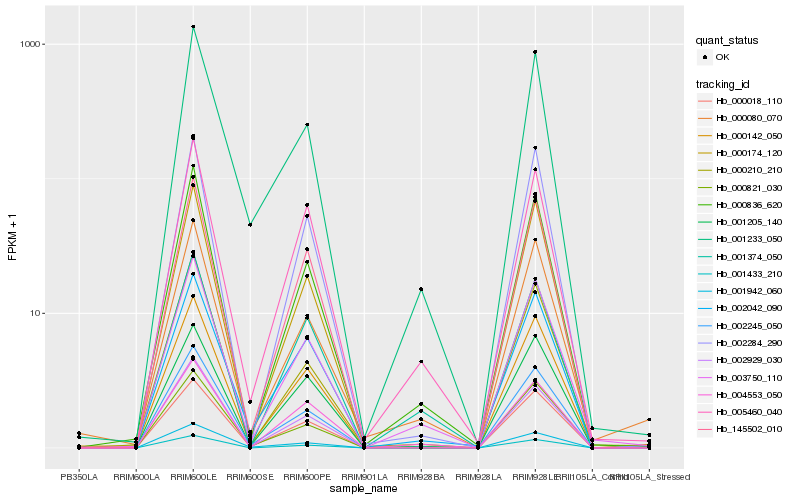
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003750_110 |
0.0 |
- |
- |
PREDICTED: aldose 1-epimerase-like [Jatropha curcas] |
| 2 |
Hb_000836_620 |
0.0541854165 |
- |
- |
PREDICTED: protochlorophyllide reductase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_005460_040 |
0.074803178 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 2 [Jatropha curcas] |
| 4 |
Hb_001374_050 |
0.0754700953 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.3 [Jatropha curcas] |
| 5 |
Hb_000142_050 |
0.0759897573 |
- |
- |
PREDICTED: beta-glucosidase 18-like isoform X1 [Populus euphratica] |
| 6 |
Hb_000080_070 |
0.0782488348 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP16-3, chloroplastic [Jatropha curcas] |
| 7 |
Hb_002042_090 |
0.0827758375 |
- |
- |
PREDICTED: root phototropism protein 3-like [Jatropha curcas] |
| 8 |
Hb_002929_030 |
0.0833482386 |
- |
- |
Serine/threonine-protein kinase-transforming protein raf, putative [Ricinus communis] |
| 9 |
Hb_001942_060 |
0.0839002238 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004553_050 |
0.0841232325 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g10020 [Jatropha curcas] |
| 11 |
Hb_000821_030 |
0.0849219033 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN-2 [Jatropha curcas] |
| 12 |
Hb_001233_050 |
0.0875504499 |
- |
- |
Magnesium-protoporphyrin IX monomethyl ester [oxidative] cyclase, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_001205_140 |
0.089853692 |
- |
- |
PREDICTED: cytochrome P450 90B1 [Jatropha curcas] |
| 14 |
Hb_002245_050 |
0.0908539931 |
- |
- |
Oxygen-evolving enhancer protein 3-1, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_000018_110 |
0.0913335202 |
- |
- |
hypothetical protein POPTR_0018s00370g [Populus trichocarpa] |
| 16 |
Hb_002284_290 |
0.0916584172 |
- |
- |
PREDICTED: transport and Golgi organization 2 homolog [Jatropha curcas] |
| 17 |
Hb_145502_010 |
0.0918842901 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB82-like [Jatropha curcas] |
| 18 |
Hb_001433_210 |
0.0919567793 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 20 [Jatropha curcas] |
| 19 |
Hb_000174_120 |
0.0920512984 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 20 |
Hb_000210_210 |
0.0934294058 |
- |
- |
PREDICTED: uncharacterized protein LOC105635822 isoform X1 [Jatropha curcas] |