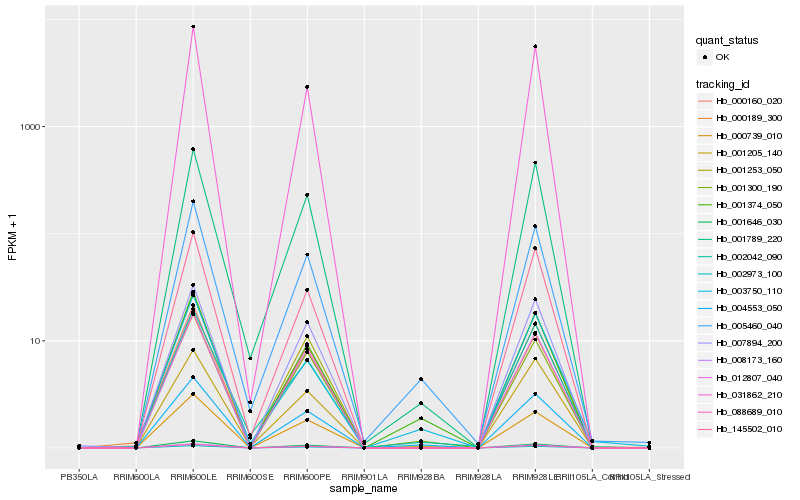
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004553_050 |
0.0 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g10020 [Jatropha curcas] |
| 2 |
Hb_001374_050 |
0.0347123673 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.3 [Jatropha curcas] |
| 3 |
Hb_005460_040 |
0.0369545251 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 2 [Jatropha curcas] |
| 4 |
Hb_001300_190 |
0.0534654345 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001789_220 |
0.0708796509 |
- |
- |
PREDICTED: BAHD acyltransferase DCR-like [Jatropha curcas] |
| 6 |
Hb_001253_050 |
0.0714150573 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 7 |
Hb_001205_140 |
0.0725084141 |
- |
- |
PREDICTED: cytochrome P450 90B1 [Jatropha curcas] |
| 8 |
Hb_002042_090 |
0.0773799836 |
- |
- |
PREDICTED: root phototropism protein 3-like [Jatropha curcas] |
| 9 |
Hb_000160_020 |
0.0786141945 |
- |
- |
PREDICTED: gamma-interferon-inducible lysosomal thiol reductase-like [Jatropha curcas] |
| 10 |
Hb_002973_100 |
0.0800785812 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable N-acetyltransferase HLS1 [Populus euphratica] |
| 11 |
Hb_001646_030 |
0.0803304986 |
- |
- |
PREDICTED: ribonuclease 3-like protein 3 [Jatropha curcas] |
| 12 |
Hb_031862_210 |
0.0803579398 |
- |
- |
hypothetical protein JCGZ_24955 [Jatropha curcas] |
| 13 |
Hb_000739_010 |
0.0805300674 |
- |
- |
PREDICTED: serine carboxypeptidase II-3-like [Vitis vinifera] |
| 14 |
Hb_012807_040 |
0.080613493 |
- |
- |
hypothetical protein POPTR_0009s01390g [Populus trichocarpa] |
| 15 |
Hb_008173_160 |
0.0811870044 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Malus domestica] |
| 16 |
Hb_088689_010 |
0.0812222544 |
- |
- |
PREDICTED: uncharacterized protein LOC105641027 [Jatropha curcas] |
| 17 |
Hb_145502_010 |
0.0821763226 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB82-like [Jatropha curcas] |
| 18 |
Hb_003750_110 |
0.0841232325 |
- |
- |
PREDICTED: aldose 1-epimerase-like [Jatropha curcas] |
| 19 |
Hb_000189_300 |
0.084538354 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 20 |
Hb_007894_200 |
0.0859961275 |
- |
- |
ubiquitin-conjugating enzyme variant, putative [Ricinus communis] |