| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
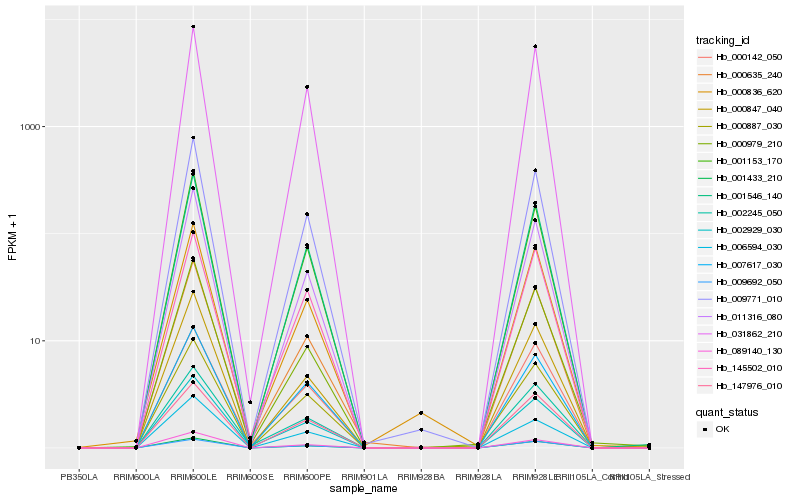
Hb_001546_140 |
0.0 |
- |
- |
Auxin-binding protein ABP19a precursor, putative [Ricinus communis] |
| 2 |
Hb_009771_010 |
0.0249408831 |
- |
- |
PREDICTED: glycerol-3-phosphate 2-O-acyltransferase 6 [Populus euphratica] |
| 3 |
Hb_007617_030 |
0.025847574 |
- |
- |
hypothetical protein JCGZ_26801 [Jatropha curcas] |
| 4 |
Hb_001153_170 |
0.0285489095 |
- |
- |
PREDICTED: protein HOTHEAD [Populus euphratica] |
| 5 |
Hb_089140_130 |
0.0327667039 |
- |
- |
hypothetical protein POPTR_0006s14510g [Populus trichocarpa] |
| 6 |
Hb_011316_080 |
0.0357792553 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 7 |
Hb_001433_210 |
0.0411143071 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 20 [Jatropha curcas] |
| 8 |
Hb_000887_030 |
0.0424719188 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 9 |
Hb_000635_240 |
0.0435382889 |
- |
- |
chlorophyll a/b-binding protein type II precursor [Populus trichocarpa] |
| 10 |
Hb_002929_030 |
0.0443578919 |
- |
- |
Serine/threonine-protein kinase-transforming protein raf, putative [Ricinus communis] |
| 11 |
Hb_031862_210 |
0.0465831669 |
- |
- |
hypothetical protein JCGZ_24955 [Jatropha curcas] |
| 12 |
Hb_006594_030 |
0.0518465947 |
- |
- |
- |
| 13 |
Hb_000847_040 |
0.0606562004 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 9 [Jatropha curcas] |
| 14 |
Hb_147976_010 |
0.0622036114 |
- |
- |
- |
| 15 |
Hb_145502_010 |
0.0624790634 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB82-like [Jatropha curcas] |
| 16 |
Hb_002245_050 |
0.063026108 |
- |
- |
Oxygen-evolving enhancer protein 3-1, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_009692_050 |
0.0646172315 |
- |
- |
- |
| 18 |
Hb_000979_210 |
0.0674938142 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000142_050 |
0.0677436897 |
- |
- |
PREDICTED: beta-glucosidase 18-like isoform X1 [Populus euphratica] |
| 20 |
Hb_000836_620 |
0.0685158732 |
- |
- |
PREDICTED: protochlorophyllide reductase, chloroplastic [Jatropha curcas] |