| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
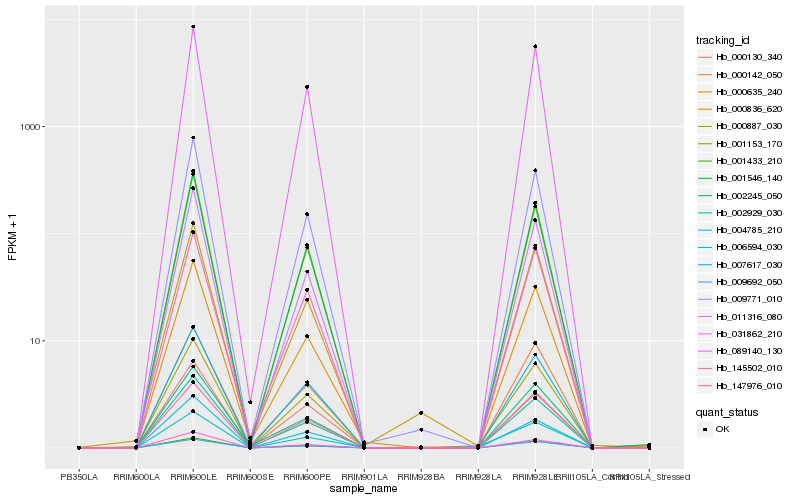
Hb_000887_030 |
0.0 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 2 |
Hb_001546_140 |
0.0424719188 |
- |
- |
Auxin-binding protein ABP19a precursor, putative [Ricinus communis] |
| 3 |
Hb_007617_030 |
0.0459849934 |
- |
- |
hypothetical protein JCGZ_26801 [Jatropha curcas] |
| 4 |
Hb_009771_010 |
0.0465687175 |
- |
- |
PREDICTED: glycerol-3-phosphate 2-O-acyltransferase 6 [Populus euphratica] |
| 5 |
Hb_001153_170 |
0.0541699004 |
- |
- |
PREDICTED: protein HOTHEAD [Populus euphratica] |
| 6 |
Hb_002929_030 |
0.0557889633 |
- |
- |
Serine/threonine-protein kinase-transforming protein raf, putative [Ricinus communis] |
| 7 |
Hb_031862_210 |
0.0559052307 |
- |
- |
hypothetical protein JCGZ_24955 [Jatropha curcas] |
| 8 |
Hb_089140_130 |
0.0563361602 |
- |
- |
hypothetical protein POPTR_0006s14510g [Populus trichocarpa] |
| 9 |
Hb_001433_210 |
0.057026414 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 20 [Jatropha curcas] |
| 10 |
Hb_011316_080 |
0.0597458029 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 11 |
Hb_000635_240 |
0.0637757583 |
- |
- |
chlorophyll a/b-binding protein type II precursor [Populus trichocarpa] |
| 12 |
Hb_006594_030 |
0.0678970284 |
- |
- |
- |
| 13 |
Hb_147976_010 |
0.068303583 |
- |
- |
- |
| 14 |
Hb_145502_010 |
0.0700053306 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB82-like [Jatropha curcas] |
| 15 |
Hb_000836_620 |
0.0706656993 |
- |
- |
PREDICTED: protochlorophyllide reductase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_009692_050 |
0.0730562442 |
- |
- |
- |
| 17 |
Hb_004785_210 |
0.0745435019 |
- |
- |
hypothetical protein JCGZ_25215 [Jatropha curcas] |
| 18 |
Hb_000130_340 |
0.0760267814 |
- |
- |
- |
| 19 |
Hb_000142_050 |
0.0763572065 |
- |
- |
PREDICTED: beta-glucosidase 18-like isoform X1 [Populus euphratica] |
| 20 |
Hb_002245_050 |
0.0777576202 |
- |
- |
Oxygen-evolving enhancer protein 3-1, chloroplast precursor, putative [Ricinus communis] |