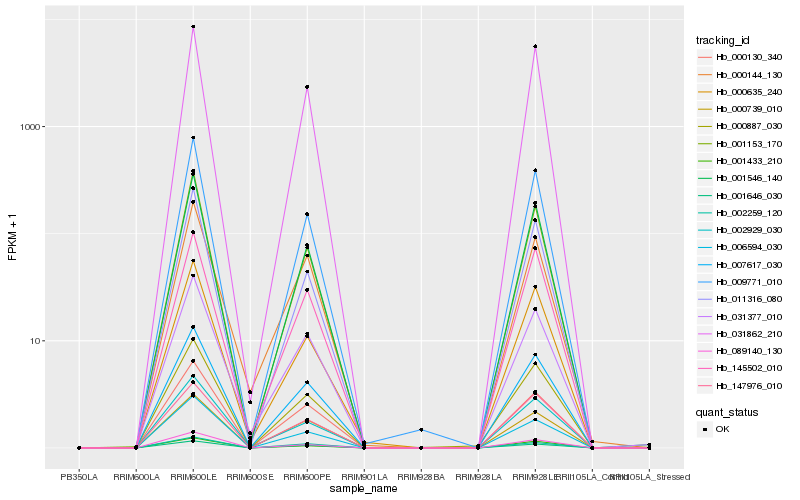
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007617_030 |
0.0 |
- |
- |
hypothetical protein JCGZ_26801 [Jatropha curcas] |
| 2 |
Hb_001546_140 |
0.025847574 |
- |
- |
Auxin-binding protein ABP19a precursor, putative [Ricinus communis] |
| 3 |
Hb_009771_010 |
0.0378927142 |
- |
- |
PREDICTED: glycerol-3-phosphate 2-O-acyltransferase 6 [Populus euphratica] |
| 4 |
Hb_001153_170 |
0.0379413464 |
- |
- |
PREDICTED: protein HOTHEAD [Populus euphratica] |
| 5 |
Hb_000887_030 |
0.0459849934 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 6 |
Hb_031862_210 |
0.0463079587 |
- |
- |
hypothetical protein JCGZ_24955 [Jatropha curcas] |
| 7 |
Hb_089140_130 |
0.0478876553 |
- |
- |
hypothetical protein POPTR_0006s14510g [Populus trichocarpa] |
| 8 |
Hb_000130_340 |
0.0484700241 |
- |
- |
- |
| 9 |
Hb_002929_030 |
0.048708207 |
- |
- |
Serine/threonine-protein kinase-transforming protein raf, putative [Ricinus communis] |
| 10 |
Hb_006594_030 |
0.0491143006 |
- |
- |
- |
| 11 |
Hb_031377_010 |
0.0542237626 |
transcription factor |
TF Family: C2C2-YABBY |
unknown [Populus trichocarpa] |
| 12 |
Hb_011316_080 |
0.056015863 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 13 |
Hb_000635_240 |
0.0581583731 |
- |
- |
chlorophyll a/b-binding protein type II precursor [Populus trichocarpa] |
| 14 |
Hb_001433_210 |
0.0599683328 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 20 [Jatropha curcas] |
| 15 |
Hb_145502_010 |
0.0617344136 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB82-like [Jatropha curcas] |
| 16 |
Hb_001646_030 |
0.0634648778 |
- |
- |
PREDICTED: ribonuclease 3-like protein 3 [Jatropha curcas] |
| 17 |
Hb_000739_010 |
0.0667309047 |
- |
- |
PREDICTED: serine carboxypeptidase II-3-like [Vitis vinifera] |
| 18 |
Hb_002259_120 |
0.0669685202 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 19 |
Hb_147976_010 |
0.0670910406 |
- |
- |
- |
| 20 |
Hb_000144_130 |
0.0689410296 |
- |
- |
conserved hypothetical protein [Ricinus communis] |