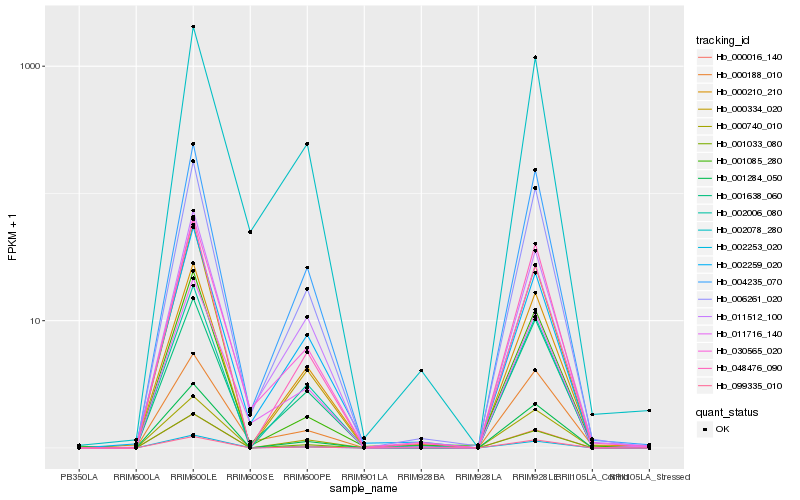
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002253_020 |
0.0 |
- |
- |
PREDICTED: callose synthase 5 [Jatropha curcas] |
| 2 |
Hb_030565_020 |
0.0573051307 |
- |
- |
PREDICTED: protein ECERIFERUM 26 [Jatropha curcas] |
| 3 |
Hb_011716_140 |
0.0585479 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein J [Jatropha curcas] |
| 4 |
Hb_000334_020 |
0.0610772213 |
- |
- |
PREDICTED: dynein light chain 2, cytoplasmic [Jatropha curcas] |
| 5 |
Hb_006261_020 |
0.0612589985 |
- |
- |
Vacuolar cation/proton exchanger 1a, putative [Ricinus communis] |
| 6 |
Hb_004235_070 |
0.0647747816 |
- |
- |
acyl carrier protein, putative [Ricinus communis] |
| 7 |
Hb_001033_080 |
0.0652359913 |
transcription factor |
TF Family: C2H2 |
Transcriptional regulator SUPERMAN, putative [Ricinus communis] |
| 8 |
Hb_000016_140 |
0.0698291095 |
- |
- |
glucose-methanol-choline (gmc) oxidoreductase, putative [Ricinus communis] |
| 9 |
Hb_002078_280 |
0.0717095535 |
- |
- |
chlorophyll a/b-binding protein type II precursor [Populus trichocarpa] |
| 10 |
Hb_001284_050 |
0.0763351355 |
- |
- |
PREDICTED: uncharacterized protein LOC100781779 isoform X1 [Glycine max] |
| 11 |
Hb_011512_100 |
0.0776401993 |
- |
- |
PREDICTED: uncharacterized protein LOC105650382 isoform X1 [Jatropha curcas] |
| 12 |
Hb_048476_090 |
0.0795891835 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002006_080 |
0.0822380284 |
- |
- |
PREDICTED: CRAL-TRIO domain-containing protein YKL091C-like isoform X2 [Citrus sinensis] |
| 14 |
Hb_001085_280 |
0.083193146 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor FAMA isoform X2 [Populus euphratica] |
| 15 |
Hb_000740_010 |
0.084075349 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 16 |
Hb_002259_020 |
0.0853070092 |
- |
- |
PREDICTED: uncharacterized protein LOC105646333 [Jatropha curcas] |
| 17 |
Hb_099335_010 |
0.0859752422 |
- |
- |
PREDICTED: UDP-glycosyltransferase 76C2-like [Populus euphratica] |
| 18 |
Hb_000188_010 |
0.0876233824 |
- |
- |
PREDICTED: cytochrome P450 94C1-like [Jatropha curcas] |
| 19 |
Hb_001638_060 |
0.089063551 |
- |
- |
PREDICTED: rho GTPase-activating protein 3-like isoform X1 [Populus euphratica] |
| 20 |
Hb_000210_210 |
0.0902553713 |
- |
- |
PREDICTED: uncharacterized protein LOC105635822 isoform X1 [Jatropha curcas] |