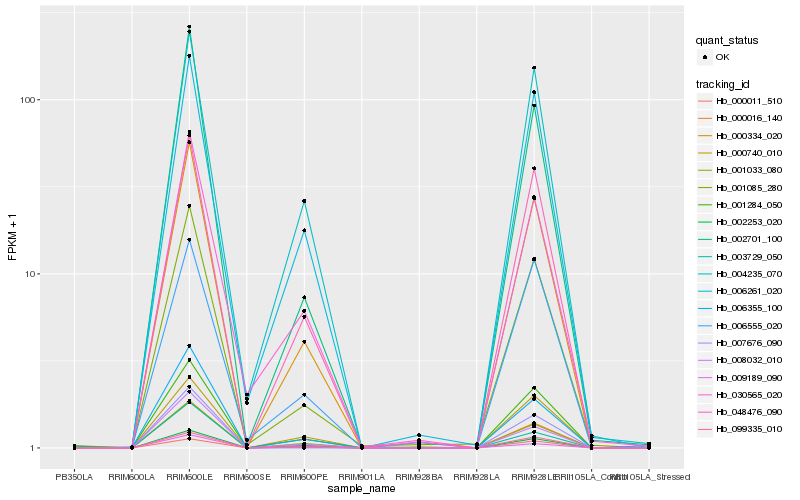
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001085_280 |
0.0 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor FAMA isoform X2 [Populus euphratica] |
| 2 |
Hb_000016_140 |
0.060159266 |
- |
- |
glucose-methanol-choline (gmc) oxidoreductase, putative [Ricinus communis] |
| 3 |
Hb_000334_020 |
0.0684825286 |
- |
- |
PREDICTED: dynein light chain 2, cytoplasmic [Jatropha curcas] |
| 4 |
Hb_001284_050 |
0.0717983209 |
- |
- |
PREDICTED: uncharacterized protein LOC100781779 isoform X1 [Glycine max] |
| 5 |
Hb_002701_100 |
0.0776310568 |
transcription factor |
TF Family: MIKC |
PREDICTED: truncated transcription factor CAULIFLOWER A-like [Jatropha curcas] |
| 6 |
Hb_002253_020 |
0.083193146 |
- |
- |
PREDICTED: callose synthase 5 [Jatropha curcas] |
| 7 |
Hb_001033_080 |
0.0856017429 |
transcription factor |
TF Family: C2H2 |
Transcriptional regulator SUPERMAN, putative [Ricinus communis] |
| 8 |
Hb_048476_090 |
0.0960336421 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_006355_100 |
0.0974973905 |
- |
- |
hypothetical protein glysoja_002455 [Glycine soja] |
| 10 |
Hb_006261_020 |
0.1001853955 |
- |
- |
Vacuolar cation/proton exchanger 1a, putative [Ricinus communis] |
| 11 |
Hb_030565_020 |
0.1029415567 |
- |
- |
PREDICTED: protein ECERIFERUM 26 [Jatropha curcas] |
| 12 |
Hb_099335_010 |
0.1043442024 |
- |
- |
PREDICTED: UDP-glycosyltransferase 76C2-like [Populus euphratica] |
| 13 |
Hb_008032_010 |
0.1109328787 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |
| 14 |
Hb_004235_070 |
0.1109402808 |
- |
- |
acyl carrier protein, putative [Ricinus communis] |
| 15 |
Hb_003729_050 |
0.1127070341 |
- |
- |
PREDICTED: vacuolar iron transporter 1-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000740_010 |
0.1167645998 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 17 |
Hb_006555_020 |
0.1173339243 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 55-like [Jatropha curcas] |
| 18 |
Hb_009189_090 |
0.1182253662 |
- |
- |
protein phosphatase-2c, putative [Ricinus communis] |
| 19 |
Hb_000011_510 |
0.1186436878 |
- |
- |
Esterase precursor, putative [Ricinus communis] |
| 20 |
Hb_007676_090 |
0.1191951035 |
- |
- |
PREDICTED: uncharacterized protein LOC105644038 [Jatropha curcas] |