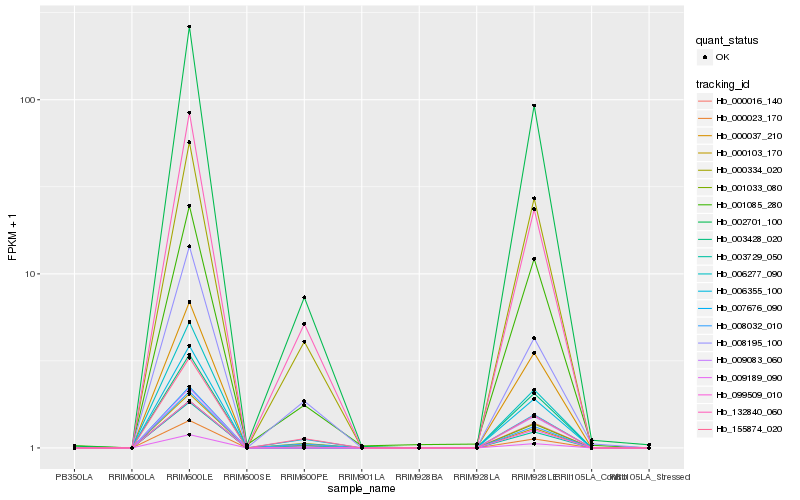
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002701_100 |
0.0 |
transcription factor |
TF Family: MIKC |
PREDICTED: truncated transcription factor CAULIFLOWER A-like [Jatropha curcas] |
| 2 |
Hb_006355_100 |
0.0448418061 |
- |
- |
hypothetical protein glysoja_002455 [Glycine soja] |
| 3 |
Hb_003729_050 |
0.0488730616 |
- |
- |
PREDICTED: vacuolar iron transporter 1-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_008032_010 |
0.0595595707 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |
| 5 |
Hb_000016_140 |
0.0599376014 |
- |
- |
glucose-methanol-choline (gmc) oxidoreductase, putative [Ricinus communis] |
| 6 |
Hb_132840_060 |
0.0721107576 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH57-like [Jatropha curcas] |
| 7 |
Hb_000334_020 |
0.0755112332 |
- |
- |
PREDICTED: dynein light chain 2, cytoplasmic [Jatropha curcas] |
| 8 |
Hb_001085_280 |
0.0776310568 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor FAMA isoform X2 [Populus euphratica] |
| 9 |
Hb_009189_090 |
0.0805585392 |
- |
- |
protein phosphatase-2c, putative [Ricinus communis] |
| 10 |
Hb_001033_080 |
0.0841558406 |
transcription factor |
TF Family: C2H2 |
Transcriptional regulator SUPERMAN, putative [Ricinus communis] |
| 11 |
Hb_000023_170 |
0.0916971098 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2-like [Jatropha curcas] |
| 12 |
Hb_155874_020 |
0.0976623136 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |
| 13 |
Hb_099509_010 |
0.0978735039 |
- |
- |
PREDICTED: vacuolar iron transporter 1-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_009083_060 |
0.1026416974 |
- |
- |
PREDICTED: uncharacterized protein LOC103344557 [Prunus mume] |
| 15 |
Hb_000037_210 |
0.102950864 |
- |
- |
- |
| 16 |
Hb_000103_170 |
0.1032007811 |
- |
- |
uncharacterized protein LOC100501254 [Zea mays] |
| 17 |
Hb_008195_100 |
0.1037220433 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: protein YABBY 4 [Vitis vinifera] |
| 18 |
Hb_003428_020 |
0.1039356158 |
- |
- |
PREDICTED: uncharacterized protein LOC105140247 [Populus euphratica] |
| 19 |
Hb_007676_090 |
0.1057171361 |
- |
- |
PREDICTED: uncharacterized protein LOC105644038 [Jatropha curcas] |
| 20 |
Hb_006277_090 |
0.1061536958 |
- |
- |
PREDICTED: uncharacterized protein LOC105649700 [Jatropha curcas] |