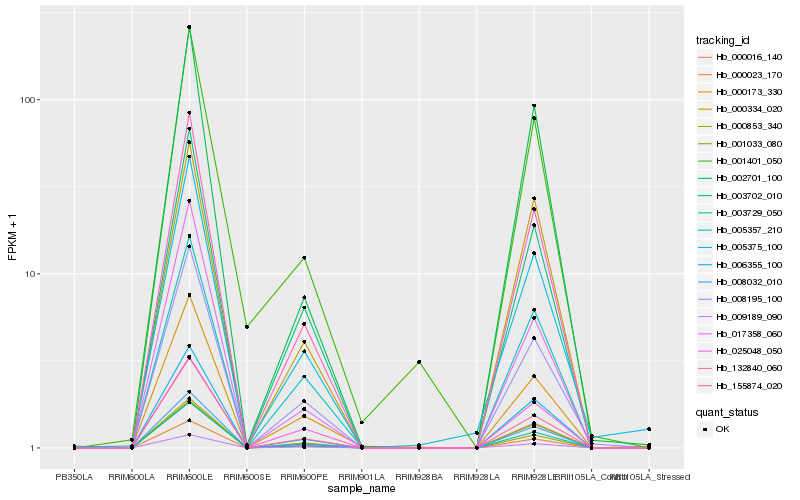
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008032_010 |
0.0 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |
| 2 |
Hb_006355_100 |
0.0171054797 |
- |
- |
hypothetical protein glysoja_002455 [Glycine soja] |
| 3 |
Hb_132840_060 |
0.0180420045 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH57-like [Jatropha curcas] |
| 4 |
Hb_003729_050 |
0.02555035 |
- |
- |
PREDICTED: vacuolar iron transporter 1-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_009189_090 |
0.0274563234 |
- |
- |
protein phosphatase-2c, putative [Ricinus communis] |
| 6 |
Hb_000023_170 |
0.0358619432 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2-like [Jatropha curcas] |
| 7 |
Hb_155874_020 |
0.0436298149 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |
| 8 |
Hb_003702_010 |
0.0521795904 |
- |
- |
PREDICTED: phospholipase A1-II 1-like [Jatropha curcas] |
| 9 |
Hb_008195_100 |
0.0583148519 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: protein YABBY 4 [Vitis vinifera] |
| 10 |
Hb_002701_100 |
0.0595595707 |
transcription factor |
TF Family: MIKC |
PREDICTED: truncated transcription factor CAULIFLOWER A-like [Jatropha curcas] |
| 11 |
Hb_005357_210 |
0.0716231214 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein MERISTEM L1 [Jatropha curcas] |
| 12 |
Hb_001033_080 |
0.0727653773 |
transcription factor |
TF Family: C2H2 |
Transcriptional regulator SUPERMAN, putative [Ricinus communis] |
| 13 |
Hb_000853_340 |
0.0768369699 |
- |
- |
PREDICTED: kynurenine--oxoglutarate transaminase 1-like isoform X2 [Populus euphratica] |
| 14 |
Hb_000173_330 |
0.0813001878 |
- |
- |
PREDICTED: glutamine synthetase leaf isozyme, chloroplastic [Vitis vinifera] |
| 15 |
Hb_000016_140 |
0.0816899472 |
- |
- |
glucose-methanol-choline (gmc) oxidoreductase, putative [Ricinus communis] |
| 16 |
Hb_005375_100 |
0.0821983133 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH57-like [Jatropha curcas] |
| 17 |
Hb_000334_020 |
0.0838771107 |
- |
- |
PREDICTED: dynein light chain 2, cytoplasmic [Jatropha curcas] |
| 18 |
Hb_017358_060 |
0.0873971606 |
- |
- |
PREDICTED: uncharacterized protein LOC105641510 [Jatropha curcas] |
| 19 |
Hb_025048_050 |
0.0920324213 |
- |
- |
PREDICTED: protein PHYTOCHROME KINASE SUBSTRATE 3 [Jatropha curcas] |
| 20 |
Hb_001401_050 |
0.0982193807 |
- |
- |
PREDICTED: chlorophyll a-b binding protein P4, chloroplastic [Jatropha curcas] |