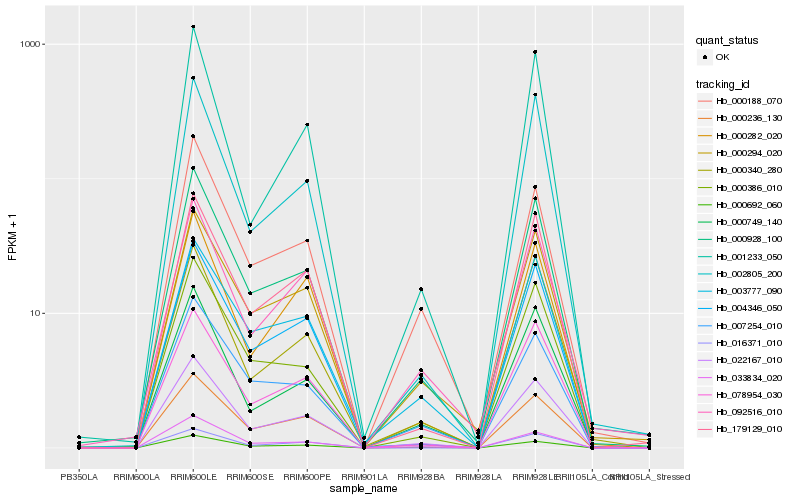
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_022167_010 |
0.0 |
- |
- |
Kinase, putative isoform 2 [Theobroma cacao] |
| 2 |
Hb_000928_100 |
0.0409250793 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 3 |
Hb_000236_130 |
0.0618959553 |
- |
- |
PREDICTED: protein ECERIFERUM 26 [Jatropha curcas] |
| 4 |
Hb_179129_010 |
0.0661884237 |
- |
- |
hypothetical protein POPTR_0018s10470g [Populus trichocarpa] |
| 5 |
Hb_000692_060 |
0.0677506207 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 6 |
Hb_033834_020 |
0.0720695365 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_004346_050 |
0.0728284509 |
- |
- |
PREDICTED: ABC transporter G family member 22 [Jatropha curcas] |
| 8 |
Hb_000749_140 |
0.0754584007 |
- |
- |
PREDICTED: 2-succinylbenzoate--CoA ligase, chloroplastic/peroxisomal [Jatropha curcas] |
| 9 |
Hb_078954_030 |
0.0758800786 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000294_020 |
0.0764559941 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 [Jatropha curcas] |
| 11 |
Hb_003777_090 |
0.0768753123 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Jatropha curcas] |
| 12 |
Hb_000386_010 |
0.0781005909 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002805_200 |
0.0831227147 |
- |
- |
PREDICTED: serine--glyoxylate aminotransferase [Nelumbo nucifera] |
| 14 |
Hb_007254_010 |
0.0842452948 |
- |
- |
hypothetical protein JCGZ_25214 [Jatropha curcas] |
| 15 |
Hb_000282_020 |
0.0845799069 |
transcription factor |
TF Family: MYB-related |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000340_280 |
0.0858418754 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000188_070 |
0.0887503195 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 18 |
Hb_016371_010 |
0.0891125061 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 19 |
Hb_092516_010 |
0.0895060196 |
- |
- |
hypothetical protein JCGZ_12248 [Jatropha curcas] |
| 20 |
Hb_001233_050 |
0.0895610856 |
- |
- |
Magnesium-protoporphyrin IX monomethyl ester [oxidative] cyclase, chloroplast precursor, putative [Ricinus communis] |