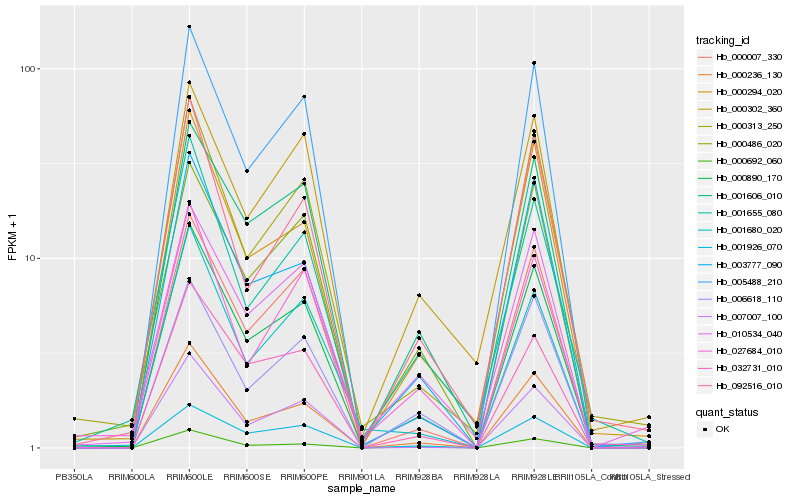
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000890_170 |
0.0 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 5 [Jatropha curcas] |
| 2 |
Hb_000236_130 |
0.0649954325 |
- |
- |
PREDICTED: protein ECERIFERUM 26 [Jatropha curcas] |
| 3 |
Hb_000313_250 |
0.0747309918 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 2 [Jatropha curcas] |
| 4 |
Hb_000007_330 |
0.0783364952 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 5 |
Hb_001926_070 |
0.0795325677 |
- |
- |
hypothetical protein JCGZ_21510 [Jatropha curcas] |
| 6 |
Hb_001606_010 |
0.0802038176 |
- |
- |
kinase, putative [Ricinus communis] |
| 7 |
Hb_000294_020 |
0.0841687912 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 [Jatropha curcas] |
| 8 |
Hb_003777_090 |
0.0886809934 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Jatropha curcas] |
| 9 |
Hb_032731_010 |
0.0887361444 |
- |
- |
RING-H2 finger protein ATL5M precursor, putative [Ricinus communis] |
| 10 |
Hb_007007_100 |
0.0900239699 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.1 [Jatropha curcas] |
| 11 |
Hb_005488_210 |
0.0932021312 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 12 |
Hb_010534_040 |
0.0950058718 |
- |
- |
PREDICTED: uncharacterized protein LOC105644385 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000486_020 |
0.0967284756 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.2 [Jatropha curcas] |
| 14 |
Hb_000692_060 |
0.0972812961 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 15 |
Hb_006618_110 |
0.0990857654 |
- |
- |
- |
| 16 |
Hb_092516_010 |
0.099680746 |
- |
- |
hypothetical protein JCGZ_12248 [Jatropha curcas] |
| 17 |
Hb_001655_080 |
0.1001725478 |
- |
- |
Purple acid phosphatase precursor, putative [Ricinus communis] |
| 18 |
Hb_000302_360 |
0.102024311 |
- |
- |
beta-carotene hydroxylase, putative [Ricinus communis] |
| 19 |
Hb_001680_020 |
0.1035778773 |
desease resistance |
Gene Name: RPW8 |
Disease resistance protein ADR1, putative [Ricinus communis] |
| 20 |
Hb_027684_010 |
0.1055429709 |
- |
- |
hypothetical protein RCOM_0504250 [Ricinus communis] |