| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
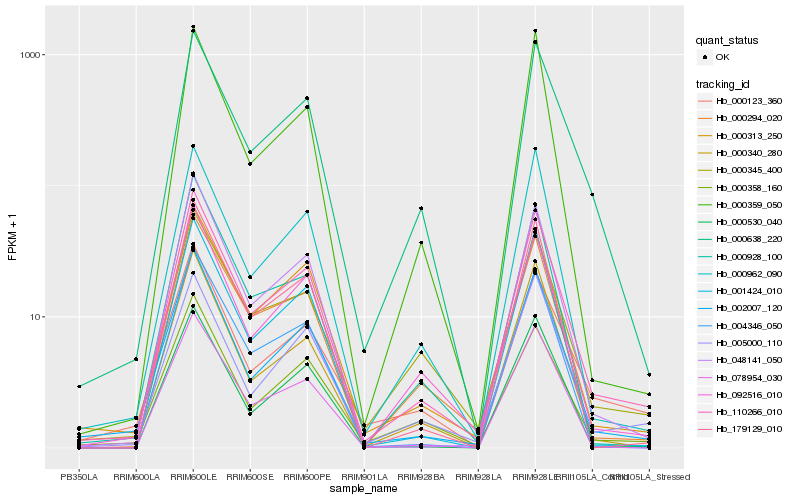
Hb_110266_010 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g21222-like [Jatropha curcas] |
| 2 |
Hb_002007_120 |
0.0613355731 |
- |
- |
PREDICTED: psbP domain-containing protein 7, chloroplastic [Jatropha curcas] |
| 3 |
Hb_092516_010 |
0.0765686342 |
- |
- |
hypothetical protein JCGZ_12248 [Jatropha curcas] |
| 4 |
Hb_000313_250 |
0.0778792913 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 2 [Jatropha curcas] |
| 5 |
Hb_048141_050 |
0.0837698206 |
- |
- |
hypothetical protein PRUPE_ppa009153mg [Prunus persica] |
| 6 |
Hb_000294_020 |
0.085055299 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 [Jatropha curcas] |
| 7 |
Hb_078954_030 |
0.0893650857 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000962_090 |
0.0903280135 |
- |
- |
omega-6 fatty acid desaturase [Hevea brasiliensis] |
| 9 |
Hb_179129_010 |
0.0909269595 |
- |
- |
hypothetical protein POPTR_0018s10470g [Populus trichocarpa] |
| 10 |
Hb_000638_220 |
0.0911313716 |
- |
- |
PREDICTED: photosystem I reaction center subunit N, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000358_160 |
0.0919186484 |
- |
- |
PREDICTED: uncharacterized protein LOC105637769 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000928_100 |
0.0945462796 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 13 |
Hb_005000_110 |
0.0945525319 |
- |
- |
PREDICTED: uncharacterized protein LOC105637872 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000345_400 |
0.0948108619 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000123_360 |
0.094864423 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 1, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_001424_010 |
0.0951905186 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 17 |
Hb_000340_280 |
0.0962806712 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000359_050 |
0.0989847961 |
- |
- |
Oxygen-evolving enhancer protein 2, chloroplast precursor, putative [Ricinus communis] |
| 19 |
Hb_000530_040 |
0.0990409573 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_004346_050 |
0.1001082143 |
- |
- |
PREDICTED: ABC transporter G family member 22 [Jatropha curcas] |