| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
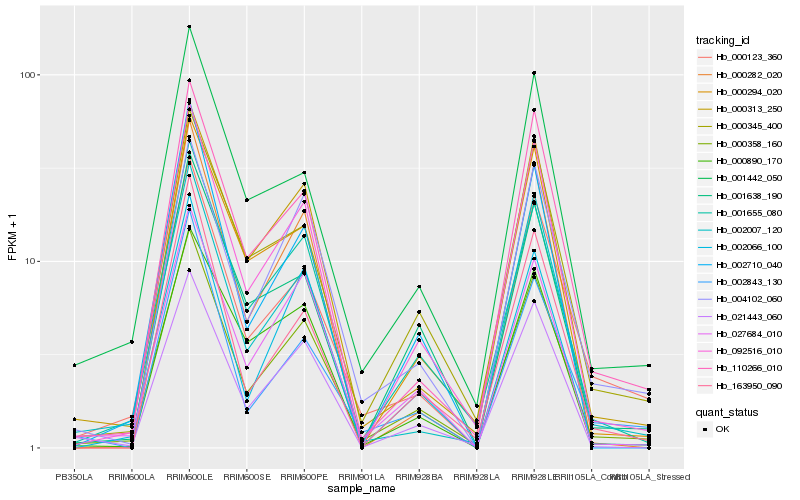
Hb_000358_160 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637769 isoform X1 [Jatropha curcas] |
| 2 |
Hb_092516_010 |
0.0614988803 |
- |
- |
hypothetical protein JCGZ_12248 [Jatropha curcas] |
| 3 |
Hb_001655_080 |
0.0693278495 |
- |
- |
Purple acid phosphatase precursor, putative [Ricinus communis] |
| 4 |
Hb_027684_010 |
0.0885448841 |
- |
- |
hypothetical protein RCOM_0504250 [Ricinus communis] |
| 5 |
Hb_021443_060 |
0.0885876444 |
- |
- |
PTH-1 family protein [Populus trichocarpa] |
| 6 |
Hb_000313_250 |
0.0888294899 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 2 [Jatropha curcas] |
| 7 |
Hb_000345_400 |
0.0914743598 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_110266_010 |
0.0919186484 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g21222-like [Jatropha curcas] |
| 9 |
Hb_000123_360 |
0.0937574086 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 1, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_002007_120 |
0.0945948235 |
- |
- |
PREDICTED: psbP domain-containing protein 7, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001442_050 |
0.0951773746 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit U, chloroplastic [Jatropha curcas] |
| 12 |
Hb_002066_100 |
0.1022469465 |
- |
- |
PREDICTED: protein disulfide-isomerase SCO2 [Jatropha curcas] |
| 13 |
Hb_001638_190 |
0.102249592 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_002710_040 |
0.1035670446 |
- |
- |
PREDICTED: cyclin-D3-3-like [Jatropha curcas] |
| 15 |
Hb_163950_090 |
0.1055146685 |
- |
- |
PREDICTED: traB domain-containing protein isoform X1 [Jatropha curcas] |
| 16 |
Hb_004102_060 |
0.1058512 |
- |
- |
JHL25H03.11 [Jatropha curcas] |
| 17 |
Hb_000294_020 |
0.1117421891 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 [Jatropha curcas] |
| 18 |
Hb_000890_170 |
0.1156766307 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 5 [Jatropha curcas] |
| 19 |
Hb_000282_020 |
0.1164782531 |
transcription factor |
TF Family: MYB-related |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002843_130 |
0.1172826347 |
- |
- |
PREDICTED: GRF1-interacting factor 2-like isoform X2 [Jatropha curcas] |