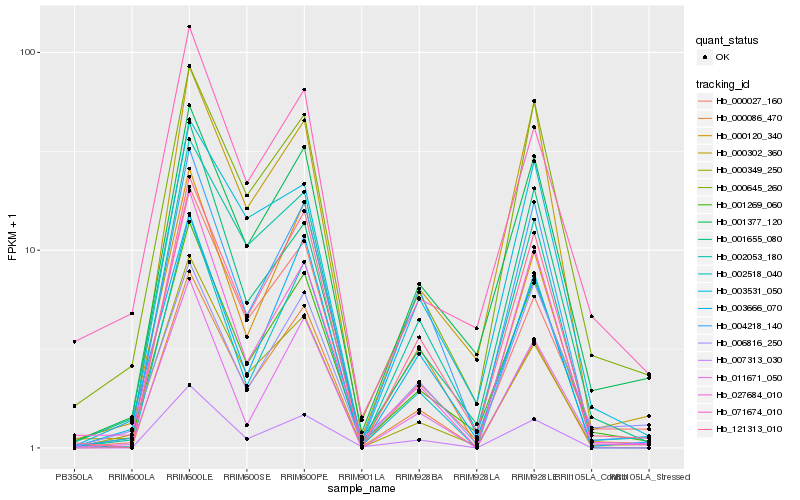
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001269_060 |
0.0 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g80640 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001377_120 |
0.06839178 |
- |
- |
PREDICTED: uncharacterized protein LOC105642861 [Jatropha curcas] |
| 3 |
Hb_121313_010 |
0.084425614 |
- |
- |
PREDICTED: endoglucanase 24-like [Jatropha curcas] |
| 4 |
Hb_000645_260 |
0.0870998884 |
- |
- |
PREDICTED: uncharacterized protein LOC105643508 [Jatropha curcas] |
| 5 |
Hb_000302_360 |
0.0886929794 |
- |
- |
beta-carotene hydroxylase, putative [Ricinus communis] |
| 6 |
Hb_004218_140 |
0.0964964439 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 isoform X3 [Jatropha curcas] |
| 7 |
Hb_027684_010 |
0.0978488586 |
- |
- |
hypothetical protein RCOM_0504250 [Ricinus communis] |
| 8 |
Hb_001655_080 |
0.0982108077 |
- |
- |
Purple acid phosphatase precursor, putative [Ricinus communis] |
| 9 |
Hb_000120_340 |
0.105346563 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 10 |
Hb_011671_050 |
0.1098183491 |
- |
- |
PREDICTED: F-box protein At5g07670 [Jatropha curcas] |
| 11 |
Hb_002518_040 |
0.1115725604 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 12 |
Hb_000349_250 |
0.1122899223 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g12460 [Jatropha curcas] |
| 13 |
Hb_000086_470 |
0.1141290339 |
- |
- |
hypothetical protein JCGZ_17645 [Jatropha curcas] |
| 14 |
Hb_003531_050 |
0.1150818169 |
- |
- |
PREDICTED: pyridoxal reductase, chloroplastic [Jatropha curcas] |
| 15 |
Hb_003666_070 |
0.1170814637 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g66560 [Jatropha curcas] |
| 16 |
Hb_071674_010 |
0.1171533302 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0006s10950g [Populus trichocarpa] |
| 17 |
Hb_007313_030 |
0.1181719438 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 [Jatropha curcas] |
| 18 |
Hb_002053_180 |
0.1182972042 |
- |
- |
PREDICTED: probable inactive receptor kinase At2g26730 [Jatropha curcas] |
| 19 |
Hb_006816_250 |
0.1185828169 |
- |
- |
PREDICTED: uncharacterized protein LOC105647444 [Jatropha curcas] |
| 20 |
Hb_000027_160 |
0.120933674 |
- |
- |
PREDICTED: probable protein phosphatase 2C 15 isoform X3 [Jatropha curcas] |