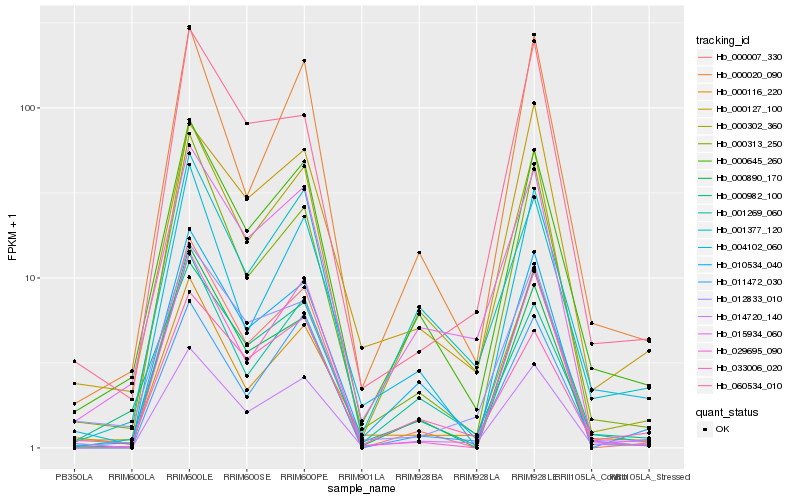
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_014720_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640367 [Jatropha curcas] |
| 2 |
Hb_000645_260 |
0.0886706521 |
- |
- |
PREDICTED: uncharacterized protein LOC105643508 [Jatropha curcas] |
| 3 |
Hb_000302_360 |
0.0906788806 |
- |
- |
beta-carotene hydroxylase, putative [Ricinus communis] |
| 4 |
Hb_010534_040 |
0.1108208856 |
- |
- |
PREDICTED: uncharacterized protein LOC105644385 isoform X1 [Jatropha curcas] |
| 5 |
Hb_012833_010 |
0.1109399868 |
- |
- |
PREDICTED: uncharacterized protein LOC105632300 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000313_250 |
0.1120657074 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 2 [Jatropha curcas] |
| 7 |
Hb_033006_020 |
0.1128597761 |
- |
- |
Aspartate aminotransferase, putative [Ricinus communis] |
| 8 |
Hb_015934_060 |
0.1144758554 |
- |
- |
PREDICTED: mitogen-activated protein kinase 20 [Jatropha curcas] |
| 9 |
Hb_011472_030 |
0.1182336476 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase [Jatropha curcas] |
| 10 |
Hb_000020_090 |
0.1197908074 |
- |
- |
hypothetical protein JCGZ_09084 [Jatropha curcas] |
| 11 |
Hb_000116_220 |
0.1224338043 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001377_120 |
0.1260321407 |
- |
- |
PREDICTED: uncharacterized protein LOC105642861 [Jatropha curcas] |
| 13 |
Hb_000127_100 |
0.126295063 |
- |
- |
PREDICTED: myb-related transcription factor, partner of profilin-like [Populus euphratica] |
| 14 |
Hb_000982_100 |
0.1266092093 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor PIF1-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_004102_060 |
0.1268071038 |
- |
- |
JHL25H03.11 [Jatropha curcas] |
| 16 |
Hb_000007_330 |
0.1272261572 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 17 |
Hb_060534_010 |
0.1274519949 |
- |
- |
diphosphoinositol polyphosphate phosphohydrolase, putative [Ricinus communis] |
| 18 |
Hb_000890_170 |
0.1277930407 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 5 [Jatropha curcas] |
| 19 |
Hb_001269_060 |
0.1282855294 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g80640 isoform X1 [Jatropha curcas] |
| 20 |
Hb_029695_090 |
0.1310452067 |
- |
- |
PREDICTED: probable inactive patatin-like protein 9 [Jatropha curcas] |