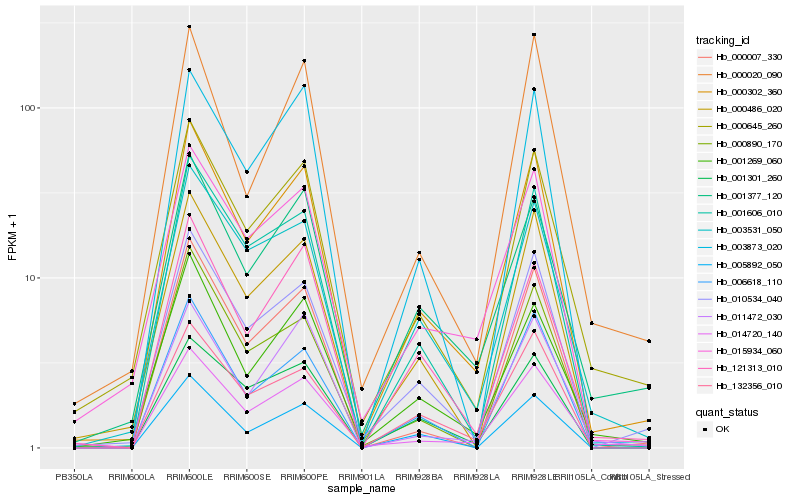
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000302_360 |
0.0 |
- |
- |
beta-carotene hydroxylase, putative [Ricinus communis] |
| 2 |
Hb_010534_040 |
0.0707752319 |
- |
- |
PREDICTED: uncharacterized protein LOC105644385 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001269_060 |
0.0886929794 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g80640 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000645_260 |
0.0887827723 |
- |
- |
PREDICTED: uncharacterized protein LOC105643508 [Jatropha curcas] |
| 5 |
Hb_001606_010 |
0.0889688367 |
- |
- |
kinase, putative [Ricinus communis] |
| 6 |
Hb_014720_140 |
0.0906788806 |
- |
- |
PREDICTED: uncharacterized protein LOC105640367 [Jatropha curcas] |
| 7 |
Hb_015934_060 |
0.0918127578 |
- |
- |
PREDICTED: mitogen-activated protein kinase 20 [Jatropha curcas] |
| 8 |
Hb_000486_020 |
0.0923263582 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.2 [Jatropha curcas] |
| 9 |
Hb_003531_050 |
0.0926267263 |
- |
- |
PREDICTED: pyridoxal reductase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_001377_120 |
0.0954889495 |
- |
- |
PREDICTED: uncharacterized protein LOC105642861 [Jatropha curcas] |
| 11 |
Hb_121313_010 |
0.0974283643 |
- |
- |
PREDICTED: endoglucanase 24-like [Jatropha curcas] |
| 12 |
Hb_132356_010 |
0.1002054056 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 13 |
Hb_000890_170 |
0.102024311 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 5 [Jatropha curcas] |
| 14 |
Hb_000007_330 |
0.1028476546 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 15 |
Hb_005892_050 |
0.1033800036 |
- |
- |
PREDICTED: uncharacterized protein LOC105632175 [Jatropha curcas] |
| 16 |
Hb_006618_110 |
0.1051229738 |
- |
- |
- |
| 17 |
Hb_003873_020 |
0.1052554669 |
- |
- |
PREDICTED: root phototropism protein 2 [Jatropha curcas] |
| 18 |
Hb_011472_030 |
0.1072349025 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase [Jatropha curcas] |
| 19 |
Hb_001301_260 |
0.1090179568 |
- |
- |
PREDICTED: 1-acyl-sn-glycerol-3-phosphate acyltransferase-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_000020_090 |
0.1092572667 |
- |
- |
hypothetical protein JCGZ_09084 [Jatropha curcas] |