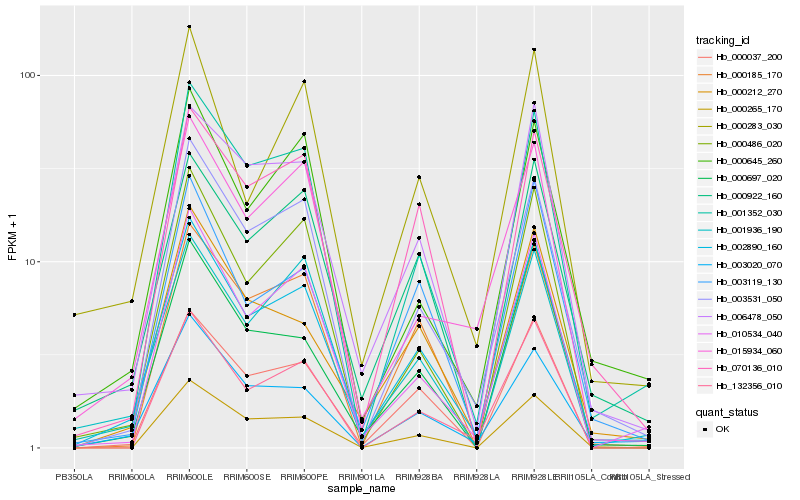
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002890_160 |
0.0 |
- |
- |
PREDICTED: protein root UVB sensitive 6 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003531_050 |
0.0893123397 |
- |
- |
PREDICTED: pyridoxal reductase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_001352_030 |
0.0903158485 |
- |
- |
PREDICTED: uncharacterized protein LOC105636310 [Jatropha curcas] |
| 4 |
Hb_003020_070 |
0.0933748925 |
- |
- |
PREDICTED: uncharacterized protein LOC103997323 [Musa acuminata subsp. malaccensis] |
| 5 |
Hb_010534_040 |
0.0937055129 |
- |
- |
PREDICTED: uncharacterized protein LOC105644385 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000037_200 |
0.0960122642 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_000922_160 |
0.0993321892 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose 5-phosphate reductoisomerase |
putative 1-deoxy-D-xylulose 5-phosphate reductoisomerase [Hevea brasiliensis] |
| 8 |
Hb_006478_050 |
0.1008967547 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000265_170 |
0.1053435505 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 10 |
Hb_132356_010 |
0.1091141181 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 11 |
Hb_000486_020 |
0.1094375661 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.2 [Jatropha curcas] |
| 12 |
Hb_000212_270 |
0.1104138564 |
- |
- |
calreticulin, putative [Ricinus communis] |
| 13 |
Hb_003119_130 |
0.1120109834 |
- |
- |
hypothetical protein POPTR_0017s07770g [Populus trichocarpa] |
| 14 |
Hb_000185_170 |
0.1131441567 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 15 |
Hb_000283_030 |
0.1136446887 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA16-like [Populus euphratica] |
| 16 |
Hb_000645_260 |
0.1156725763 |
- |
- |
PREDICTED: uncharacterized protein LOC105643508 [Jatropha curcas] |
| 17 |
Hb_000697_020 |
0.1164056119 |
- |
- |
Zinc-binding dehydrogenase family protein isoform 1 [Theobroma cacao] |
| 18 |
Hb_070136_010 |
0.1169806078 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 19 |
Hb_001936_190 |
0.1178451326 |
- |
- |
hypothetical protein JCGZ_17780 [Jatropha curcas] |
| 20 |
Hb_015934_060 |
0.1190019458 |
- |
- |
PREDICTED: mitogen-activated protein kinase 20 [Jatropha curcas] |