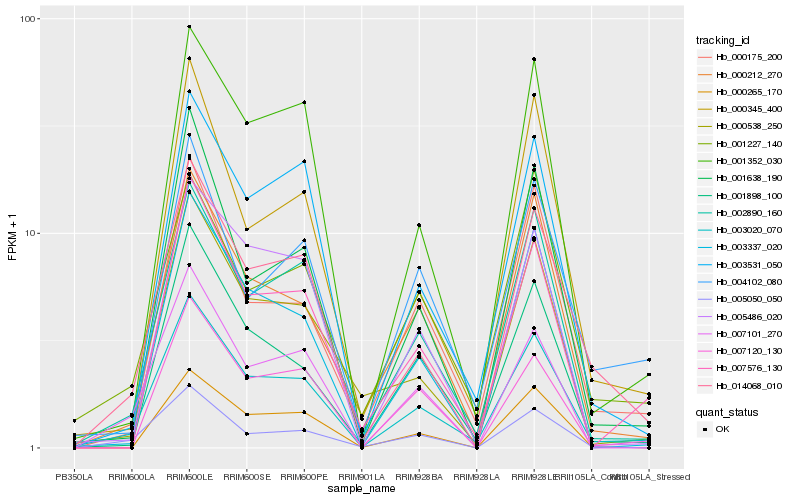
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003020_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103997323 [Musa acuminata subsp. malaccensis] |
| 2 |
Hb_002890_160 |
0.0933748925 |
- |
- |
PREDICTED: protein root UVB sensitive 6 isoform X1 [Jatropha curcas] |
| 3 |
Hb_014068_010 |
0.1069950929 |
- |
- |
PREDICTED: phototropin-1 [Jatropha curcas] |
| 4 |
Hb_000212_270 |
0.1143678603 |
- |
- |
calreticulin, putative [Ricinus communis] |
| 5 |
Hb_003531_050 |
0.1218930387 |
- |
- |
PREDICTED: pyridoxal reductase, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001352_030 |
0.1225233798 |
- |
- |
PREDICTED: uncharacterized protein LOC105636310 [Jatropha curcas] |
| 7 |
Hb_001898_100 |
0.1239221429 |
- |
- |
PREDICTED: phosphatidylserine decarboxylase proenzyme 2-like [Jatropha curcas] |
| 8 |
Hb_000175_200 |
0.125625655 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000345_400 |
0.1311564026 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001638_190 |
0.1312916167 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_003337_020 |
0.1324169177 |
- |
- |
PREDICTED: uncharacterized protein LOC105116455 [Populus euphratica] |
| 12 |
Hb_007576_130 |
0.1326335755 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At5g64080 [Jatropha curcas] |
| 13 |
Hb_005050_050 |
0.1345143853 |
- |
- |
PREDICTED: wall-associated receptor kinase 5-like [Glycine max] |
| 14 |
Hb_000538_250 |
0.1347548377 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g07650 [Jatropha curcas] |
| 15 |
Hb_007101_270 |
0.1353701464 |
- |
- |
PREDICTED: uncharacterized protein LOC105640496 [Jatropha curcas] |
| 16 |
Hb_001227_140 |
0.13596064 |
- |
- |
PREDICTED: protein ROS1 [Jatropha curcas] |
| 17 |
Hb_005486_020 |
0.1360576339 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT5 [Jatropha curcas] |
| 18 |
Hb_004102_080 |
0.1391170779 |
- |
- |
PREDICTED: acyl-protein thioesterase 2 [Jatropha curcas] |
| 19 |
Hb_000265_170 |
0.1407609107 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 20 |
Hb_007120_130 |
0.1471279623 |
- |
- |
conserved hypothetical protein [Ricinus communis] |