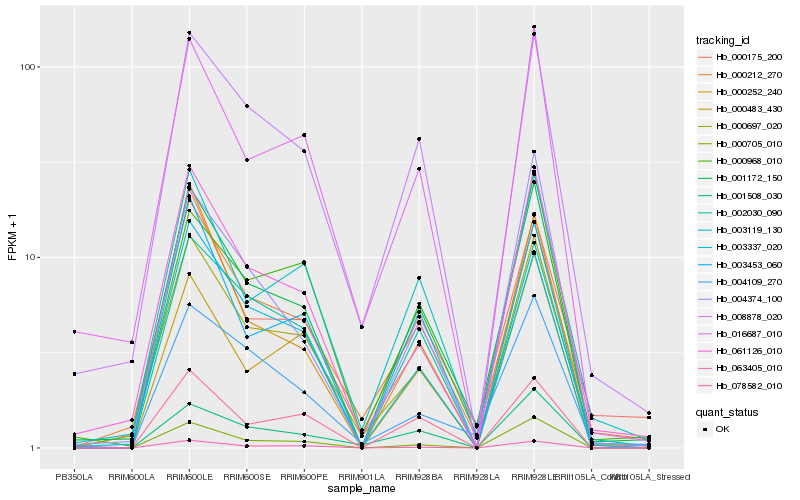
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001172_150 |
0.0 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: putative ABC transporter C family member 15 [Jatropha curcas] |
| 2 |
Hb_000697_020 |
0.0892962513 |
- |
- |
Zinc-binding dehydrogenase family protein isoform 1 [Theobroma cacao] |
| 3 |
Hb_003119_130 |
0.0992631085 |
- |
- |
hypothetical protein POPTR_0017s07770g [Populus trichocarpa] |
| 4 |
Hb_008878_020 |
0.0994992077 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 3 member H1-like [Populus euphratica] |
| 5 |
Hb_000212_270 |
0.1008197418 |
- |
- |
calreticulin, putative [Ricinus communis] |
| 6 |
Hb_061126_010 |
0.1036068638 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 7 |
Hb_000705_010 |
0.1114604915 |
- |
- |
hypothetical protein CICLE_v10027296mg [Citrus clementina] |
| 8 |
Hb_002030_090 |
0.1119681424 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 9 |
Hb_016687_010 |
0.1129595754 |
- |
- |
PREDICTED: uncharacterized protein LOC105637401 [Jatropha curcas] |
| 10 |
Hb_000175_200 |
0.1148869405 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003453_060 |
0.1213471376 |
- |
- |
PREDICTED: nudix hydrolase 13, mitochondrial [Jatropha curcas] |
| 12 |
Hb_000968_010 |
0.1226797915 |
- |
- |
PREDICTED: glutamate receptor 3.7 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003337_020 |
0.1231235064 |
- |
- |
PREDICTED: uncharacterized protein LOC105116455 [Populus euphratica] |
| 14 |
Hb_004109_270 |
0.1233818735 |
transcription factor |
TF Family: NAC |
transcription activator NAC1 family protein [Populus trichocarpa] |
| 15 |
Hb_063405_010 |
0.1243580697 |
- |
- |
PREDICTED: probable terpene synthase 3 [Jatropha curcas] |
| 16 |
Hb_004374_100 |
0.1252149068 |
- |
- |
PREDICTED: phosphoglycerate mutase-like protein AT74H [Jatropha curcas] |
| 17 |
Hb_078582_010 |
0.1254010071 |
- |
- |
hypothetical protein POPTR_0005s01620g [Populus trichocarpa] |
| 18 |
Hb_000252_240 |
0.1297605855 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like isoform X2 [Gossypium raimondii] |
| 19 |
Hb_000483_430 |
0.1311890804 |
- |
- |
- |
| 20 |
Hb_001508_030 |
0.132360805 |
- |
- |
hypothetical protein VITISV_029808 [Vitis vinifera] |