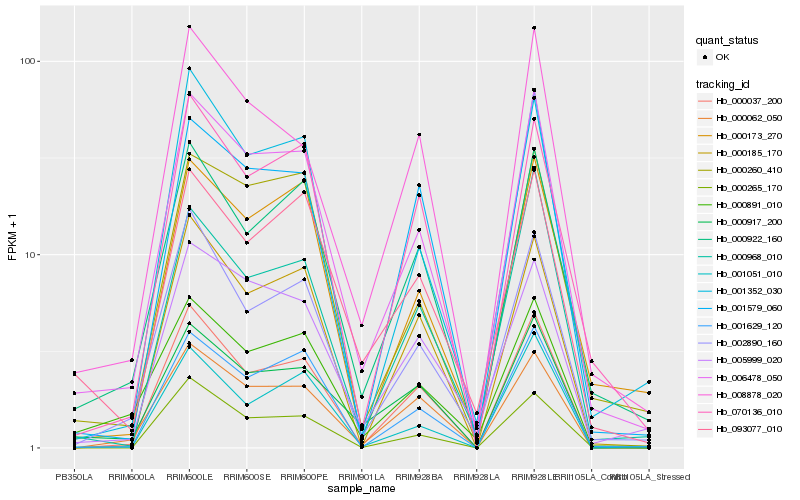
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006478_050 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001629_120 |
0.0945193298 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At4g27220 isoform X2 [Populus euphratica] |
| 3 |
Hb_000922_160 |
0.098742055 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose 5-phosphate reductoisomerase |
putative 1-deoxy-D-xylulose 5-phosphate reductoisomerase [Hevea brasiliensis] |
| 4 |
Hb_000037_200 |
0.1005679153 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_002890_160 |
0.1008967547 |
- |
- |
PREDICTED: protein root UVB sensitive 6 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000260_410 |
0.1025482354 |
- |
- |
PREDICTED: protein LURP-one-related 8 [Jatropha curcas] |
| 7 |
Hb_001352_030 |
0.1077507059 |
- |
- |
PREDICTED: uncharacterized protein LOC105636310 [Jatropha curcas] |
| 8 |
Hb_000891_010 |
0.1082690562 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_005999_020 |
0.1098937076 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1-like [Jatropha curcas] |
| 10 |
Hb_008878_020 |
0.1115934406 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 3 member H1-like [Populus euphratica] |
| 11 |
Hb_000173_270 |
0.1120574914 |
- |
- |
PREDICTED: mitogen-activated protein kinase 15 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000265_170 |
0.1124864302 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 13 |
Hb_093077_010 |
0.1140953779 |
transcription factor |
TF Family: G2-like |
PREDICTED: two-component response regulator-like APRR2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000917_200 |
0.1160246511 |
- |
- |
hypothetical protein PRUPE_ppa010555mg [Prunus persica] |
| 15 |
Hb_000062_050 |
0.116355128 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 16 |
Hb_000968_010 |
0.116979813 |
- |
- |
PREDICTED: glutamate receptor 3.7 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000185_170 |
0.1174353613 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 18 |
Hb_070136_010 |
0.1189159533 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 19 |
Hb_001579_060 |
0.1200407894 |
- |
- |
carbonic anhydrase, putative [Ricinus communis] |
| 20 |
Hb_001051_010 |
0.1205300066 |
- |
- |
PREDICTED: putative cyclic nucleotide-gated ion channel 15 [Jatropha curcas] |