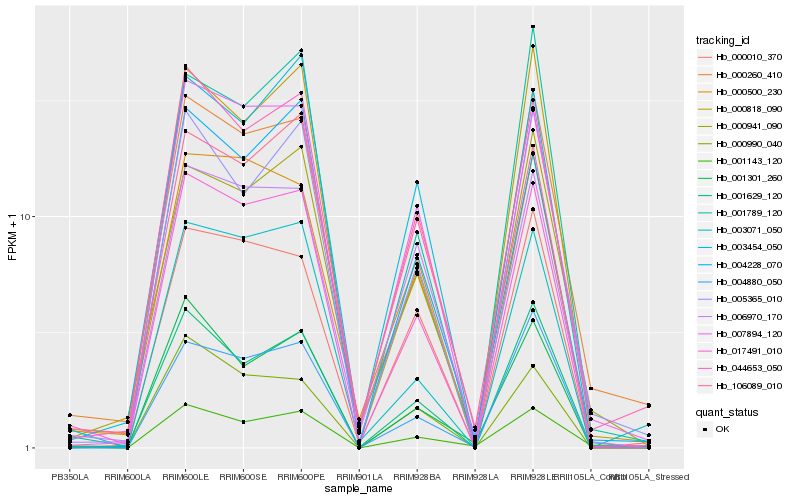
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_017491_010 |
0.0 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_007894_120 |
0.0756979953 |
- |
- |
PREDICTED: 1-acyl-sn-glycerol-3-phosphate acyltransferase-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_044653_050 |
0.0811184664 |
- |
- |
PREDICTED: CTL-like protein DDB_G0274487 isoform X1 [Jatropha curcas] |
| 4 |
Hb_003071_050 |
0.0831644687 |
transcription factor |
TF Family: ERF |
ethylene-responsive element binding protein 2 [Hevea brasiliensis] |
| 5 |
Hb_000260_410 |
0.0840345279 |
- |
- |
PREDICTED: protein LURP-one-related 8 [Jatropha curcas] |
| 6 |
Hb_000500_230 |
0.0884256695 |
- |
- |
hypothetical protein RCOM_0634810 [Ricinus communis] |
| 7 |
Hb_000818_090 |
0.0999400116 |
- |
- |
glutamate receptor 3 plant, putative [Ricinus communis] |
| 8 |
Hb_006970_170 |
0.1008370427 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_001629_120 |
0.1027521241 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At4g27220 isoform X2 [Populus euphratica] |
| 10 |
Hb_003454_050 |
0.1072879762 |
- |
- |
PREDICTED: probable methyltransferase PMT14 [Jatropha curcas] |
| 11 |
Hb_000990_040 |
0.1073825471 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 7 [Jatropha curcas] |
| 12 |
Hb_005365_010 |
0.1076699878 |
- |
- |
PREDICTED: serine carboxypeptidase-like 20 [Jatropha curcas] |
| 13 |
Hb_000010_370 |
0.1077894301 |
- |
- |
PREDICTED: uncharacterized protein LOC105640263 [Jatropha curcas] |
| 14 |
Hb_004880_050 |
0.1122999985 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000941_090 |
0.1124677394 |
- |
- |
PREDICTED: protein trichome birefringence [Jatropha curcas] |
| 16 |
Hb_004228_070 |
0.1126110938 |
- |
- |
PREDICTED: uncharacterized protein LOC103338699 [Prunus mume] |
| 17 |
Hb_106089_010 |
0.114020433 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 18 |
Hb_001301_260 |
0.114222583 |
- |
- |
PREDICTED: 1-acyl-sn-glycerol-3-phosphate acyltransferase-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_001143_120 |
0.1174619998 |
- |
- |
PREDICTED: uncharacterized protein LOC105638955 [Jatropha curcas] |
| 20 |
Hb_001789_120 |
0.1178731008 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic-like [Jatropha curcas] |