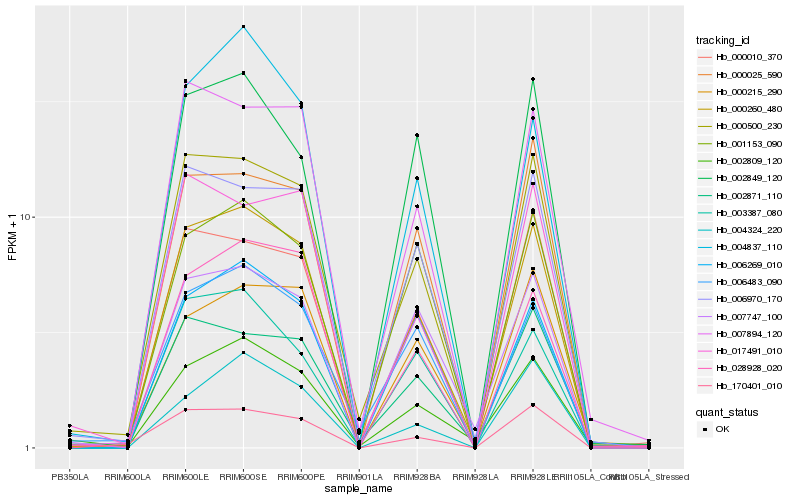
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000260_480 |
0.0 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SCARECROW [Jatropha curcas] |
| 2 |
Hb_000500_230 |
0.0742775895 |
- |
- |
hypothetical protein RCOM_0634810 [Ricinus communis] |
| 3 |
Hb_000010_370 |
0.0947961785 |
- |
- |
PREDICTED: uncharacterized protein LOC105640263 [Jatropha curcas] |
| 4 |
Hb_006970_170 |
0.0949541114 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_007894_120 |
0.0963927422 |
- |
- |
PREDICTED: 1-acyl-sn-glycerol-3-phosphate acyltransferase-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_028928_020 |
0.1058561779 |
- |
- |
hypothetical protein CICLE_v10018504mg [Citrus clementina] |
| 7 |
Hb_000025_590 |
0.1096499994 |
- |
- |
PREDICTED: cytochrome P450 94A1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_002871_110 |
0.1110768683 |
transcription factor |
TF Family: ARF |
hypothetical protein POPTR_0014s12980g [Populus trichocarpa] |
| 9 |
Hb_002809_120 |
0.1119821486 |
- |
- |
Uncharacterized protein TCM_031281 [Theobroma cacao] |
| 10 |
Hb_006269_010 |
0.1121495121 |
- |
- |
hypothetical protein JCGZ_21552 [Jatropha curcas] |
| 11 |
Hb_007747_100 |
0.1136303884 |
- |
- |
PREDICTED: probable F-box protein At1g60180 [Jatropha curcas] |
| 12 |
Hb_004837_110 |
0.1142222547 |
- |
- |
PREDICTED: MATE efflux family protein 1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_170401_010 |
0.1142383702 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 14 |
Hb_001153_090 |
0.1182101731 |
- |
- |
PREDICTED: protein FAM63A [Jatropha curcas] |
| 15 |
Hb_002849_120 |
0.1191585487 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 16 |
Hb_003387_080 |
0.1194211878 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 17 |
Hb_000215_290 |
0.1194448317 |
transcription factor |
TF Family: NAC |
PREDICTED: protein BEARSKIN2 isoform X2 [Jatropha curcas] |
| 18 |
Hb_006483_090 |
0.1202059305 |
- |
- |
PREDICTED: uncharacterized protein LOC105639979 [Jatropha curcas] |
| 19 |
Hb_017491_010 |
0.1208368628 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_004324_220 |
0.1213948765 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 53 [Jatropha curcas] |