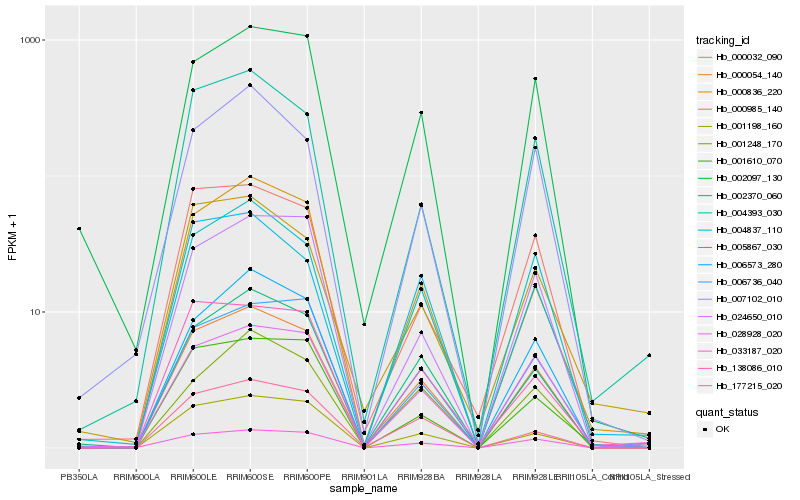
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000054_140 |
0.0 |
- |
- |
PREDICTED: MACPF domain-containing protein CAD1 [Jatropha curcas] |
| 2 |
Hb_002370_060 |
0.0737490576 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Populus euphratica] |
| 3 |
Hb_006573_280 |
0.0825242045 |
- |
- |
PREDICTED: probable membrane-associated kinase regulator 5 [Jatropha curcas] |
| 4 |
Hb_004837_110 |
0.0856727363 |
- |
- |
PREDICTED: MATE efflux family protein 1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001198_160 |
0.0919222604 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 6 |
Hb_000836_220 |
0.0921648639 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 7 |
Hb_033187_020 |
0.0937474022 |
- |
- |
hypothetical protein CARUB_v10004250mg, partial [Capsella rubella] |
| 8 |
Hb_004393_030 |
0.0988335818 |
- |
- |
CBL-interacting protein kinase 6 [Populus trichocarpa] |
| 9 |
Hb_028928_020 |
0.1008532163 |
- |
- |
hypothetical protein CICLE_v10018504mg [Citrus clementina] |
| 10 |
Hb_177215_020 |
0.1036468205 |
- |
- |
acc synthase, putative [Ricinus communis] |
| 11 |
Hb_002097_130 |
0.1121458869 |
- |
- |
tonoplast intrinsic protein [Hevea brasiliensis] |
| 12 |
Hb_001248_170 |
0.1135444667 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 13 |
Hb_005867_030 |
0.1144440284 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |
| 14 |
Hb_007102_010 |
0.11460192 |
- |
- |
Granule-bound starch synthase 1, chloroplastic/amyloplastic [Gossypium arboreum] |
| 15 |
Hb_000032_090 |
0.1169915049 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_024650_010 |
0.1170885938 |
- |
- |
PREDICTED: BAHD acyltransferase DCR [Jatropha curcas] |
| 17 |
Hb_006736_040 |
0.1179174538 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase, basic isoform-like [Populus euphratica] |
| 18 |
Hb_138086_010 |
0.1191816322 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 19 |
Hb_000985_140 |
0.1215206363 |
- |
- |
PREDICTED: uncharacterized protein LOC105643381 isoform X3 [Jatropha curcas] |
| 20 |
Hb_001610_070 |
0.1220370304 |
- |
- |
hypothetical protein MIMGU_mgv1a022867mg [Erythranthe guttata] |