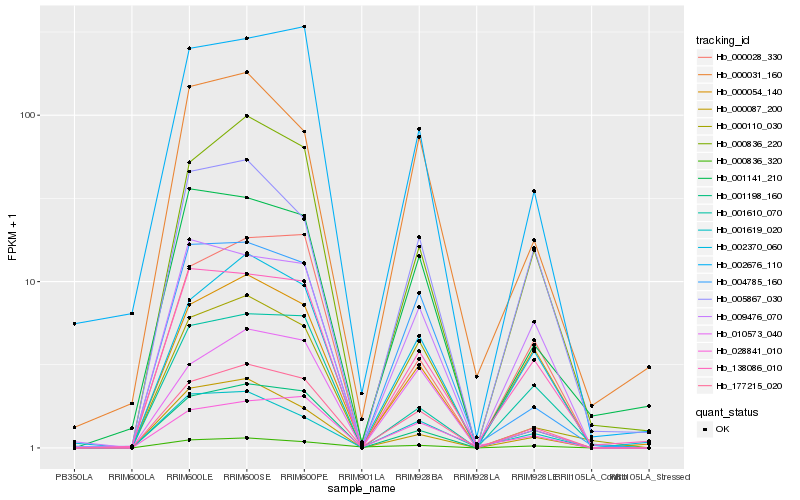
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_177215_020 |
0.0 |
- |
- |
acc synthase, putative [Ricinus communis] |
| 2 |
Hb_001198_160 |
0.0782049558 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 3 |
Hb_002370_060 |
0.0870348401 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Populus euphratica] |
| 4 |
Hb_000836_220 |
0.0926809039 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 5 |
Hb_000054_140 |
0.1036468205 |
- |
- |
PREDICTED: MACPF domain-containing protein CAD1 [Jatropha curcas] |
| 6 |
Hb_138086_010 |
0.1106800448 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 7 |
Hb_028841_010 |
0.1135760599 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase CES101 [Jatropha curcas] |
| 8 |
Hb_010573_040 |
0.1142173648 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal isoform X2 [Populus euphratica] |
| 9 |
Hb_002676_110 |
0.1202330655 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase 2-like [Gossypium raimondii] |
| 10 |
Hb_000836_320 |
0.1234002869 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 11 |
Hb_004785_160 |
0.126183462 |
- |
- |
PREDICTED: U-box domain-containing protein 28-like [Jatropha curcas] |
| 12 |
Hb_000028_330 |
0.1276758564 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 9-like [Pyrus x bretschneideri] |
| 13 |
Hb_001619_020 |
0.1282300598 |
- |
- |
LRR receptor-like kinase family protein [Medicago truncatula] |
| 14 |
Hb_000110_030 |
0.1287498125 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 15 |
Hb_009476_070 |
0.1294991897 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 2 [Jatropha curcas] |
| 16 |
Hb_001610_070 |
0.1348111653 |
- |
- |
hypothetical protein MIMGU_mgv1a022867mg [Erythranthe guttata] |
| 17 |
Hb_001141_210 |
0.1356136372 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105632009 [Jatropha curcas] |
| 18 |
Hb_000087_200 |
0.1381743436 |
- |
- |
PREDICTED: uncharacterized protein LOC105641954 [Jatropha curcas] |
| 19 |
Hb_000031_160 |
0.1415002902 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 17 [Jatropha curcas] |
| 20 |
Hb_005867_030 |
0.1428690126 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |