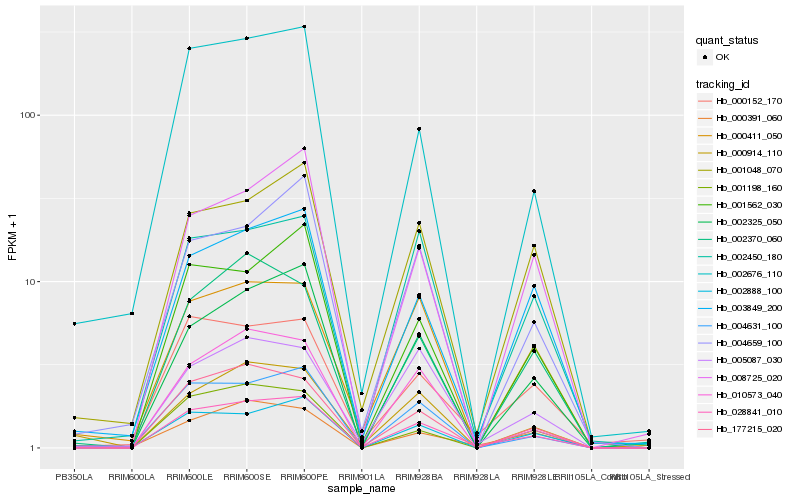
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028841_010 |
0.0 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase CES101 [Jatropha curcas] |
| 2 |
Hb_177215_020 |
0.1135760599 |
- |
- |
acc synthase, putative [Ricinus communis] |
| 3 |
Hb_004631_100 |
0.117436058 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH96 [Jatropha curcas] |
| 4 |
Hb_002325_050 |
0.1212215361 |
transcription factor |
TF Family: MYB |
MYB transcriptional factor [Populus nigra] |
| 5 |
Hb_002676_110 |
0.1217131099 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase 2-like [Gossypium raimondii] |
| 6 |
Hb_003849_200 |
0.1235534396 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g67900-like isoform X1 [Citrus sinensis] |
| 7 |
Hb_002888_100 |
0.1241931467 |
- |
- |
PREDICTED: uncharacterized protein LOC103699074, partial [Phoenix dactylifera] |
| 8 |
Hb_000152_170 |
0.1246853677 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 homolog [Jatropha curcas] |
| 9 |
Hb_001562_030 |
0.1301099374 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH35 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001048_070 |
0.1308269141 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0007s14590g [Populus trichocarpa] |
| 11 |
Hb_010573_040 |
0.1355922312 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal isoform X2 [Populus euphratica] |
| 12 |
Hb_000391_060 |
0.136520785 |
- |
- |
PREDICTED: uncharacterized protein LOC104445137 [Eucalyptus grandis] |
| 13 |
Hb_002450_180 |
0.1384744053 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001198_160 |
0.138694725 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 15 |
Hb_004659_100 |
0.1399801219 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002370_060 |
0.1411164475 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Populus euphratica] |
| 17 |
Hb_000914_110 |
0.1412945459 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein WIP2 [Jatropha curcas] |
| 18 |
Hb_008725_020 |
0.1422909694 |
- |
- |
PREDICTED: cytidine deaminase 1 [Jatropha curcas] |
| 19 |
Hb_005087_030 |
0.1424679738 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |
| 20 |
Hb_000411_050 |
0.1439172436 |
- |
- |
hypothetical protein AMTR_s00066p00166230 [Amborella trichopoda] |