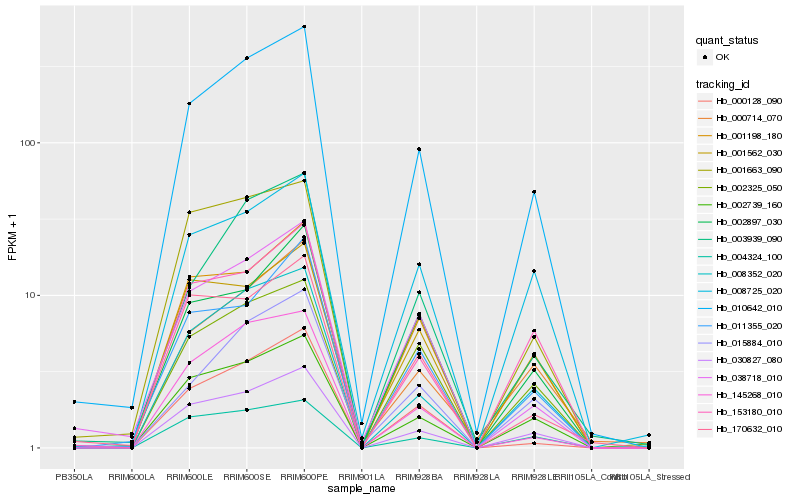
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010642_010 |
0.0 |
- |
- |
PREDICTED: aquaporin TIP2-1 [Jatropha curcas] |
| 2 |
Hb_030827_080 |
0.0637584216 |
- |
- |
PREDICTED: uncharacterized protein LOC105648491 [Jatropha curcas] |
| 3 |
Hb_002739_160 |
0.0701414296 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 4 |
Hb_008352_020 |
0.0750932093 |
- |
- |
hypothetical protein [Cecembia lonarensis] |
| 5 |
Hb_038718_010 |
0.0955284764 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 6 |
Hb_003939_090 |
0.0969716523 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 7 |
Hb_001198_180 |
0.0992924696 |
- |
- |
RecName: Full=Anthocyanidin 3-O-glucosyltransferase 1; AltName: Full=Flavonol 3-O-glucosyltransferase 1; AltName: Full=UDP-glucose flavonoid 3-O-glucosyltransferase 1 [Manihot esculenta] |
| 8 |
Hb_015884_010 |
0.0996008421 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X1 [Jatropha curcas] |
| 9 |
Hb_145268_010 |
0.1037432361 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Jatropha curcas] |
| 10 |
Hb_000128_090 |
0.1042088967 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 11 |
Hb_002325_050 |
0.1052756992 |
transcription factor |
TF Family: MYB |
MYB transcriptional factor [Populus nigra] |
| 12 |
Hb_000714_070 |
0.1070595522 |
transcription factor |
TF Family: LOB |
hypothetical protein JCGZ_14754 [Jatropha curcas] |
| 13 |
Hb_153180_010 |
0.1137343312 |
- |
- |
NBS-LRR disease resistance protein NBS49 [Dimocarpus longan] |
| 14 |
Hb_011355_020 |
0.1148926709 |
- |
- |
extensin, proline-rich protein, putative [Ricinus communis] |
| 15 |
Hb_001663_090 |
0.1178096481 |
- |
- |
ribonuclease t2, putative [Ricinus communis] |
| 16 |
Hb_008725_020 |
0.1202453183 |
- |
- |
PREDICTED: cytidine deaminase 1 [Jatropha curcas] |
| 17 |
Hb_002897_030 |
0.121125793 |
- |
- |
PREDICTED: methylesterase 17 [Jatropha curcas] |
| 18 |
Hb_170632_010 |
0.1251330902 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 19 |
Hb_001562_030 |
0.1313452467 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH35 isoform X1 [Jatropha curcas] |
| 20 |
Hb_004324_100 |
0.1314011308 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |