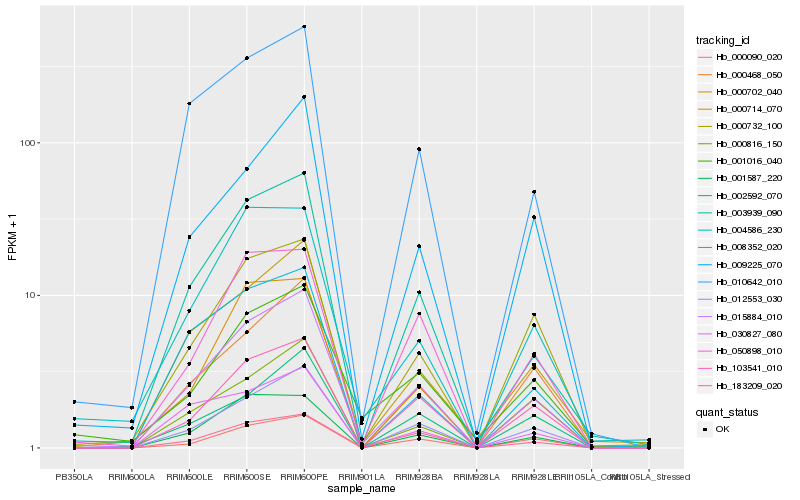
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015884_010 |
0.0 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X1 [Jatropha curcas] |
| 2 |
Hb_012553_030 |
0.0784952908 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 3 |
Hb_003939_090 |
0.0899552494 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 4 |
Hb_010642_010 |
0.0996008421 |
- |
- |
PREDICTED: aquaporin TIP2-1 [Jatropha curcas] |
| 5 |
Hb_000714_070 |
0.1054711045 |
transcription factor |
TF Family: LOB |
hypothetical protein JCGZ_14754 [Jatropha curcas] |
| 6 |
Hb_103541_010 |
0.1117780704 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000468_050 |
0.1151322984 |
- |
- |
- |
| 8 |
Hb_000090_020 |
0.1195857963 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 9 |
Hb_009225_070 |
0.1206427537 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 10 |
Hb_000732_100 |
0.121693174 |
- |
- |
PREDICTED: uncharacterized protein LOC105649440 [Jatropha curcas] |
| 11 |
Hb_183209_020 |
0.1240426438 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X2 [Jatropha curcas] |
| 12 |
Hb_050898_010 |
0.1286875203 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 13 |
Hb_000702_040 |
0.1289143929 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 14 |
Hb_001587_220 |
0.1291321945 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 15 |
Hb_008352_020 |
0.132857991 |
- |
- |
hypothetical protein [Cecembia lonarensis] |
| 16 |
Hb_001016_040 |
0.1337994266 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002592_070 |
0.1338124868 |
- |
- |
hypothetical protein VITISV_020993 [Vitis vinifera] |
| 18 |
Hb_000816_150 |
0.1364222369 |
- |
- |
oligopeptide transporter, putative [Ricinus communis] |
| 19 |
Hb_004586_230 |
0.1372888461 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_030827_080 |
0.1383758399 |
- |
- |
PREDICTED: uncharacterized protein LOC105648491 [Jatropha curcas] |