| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
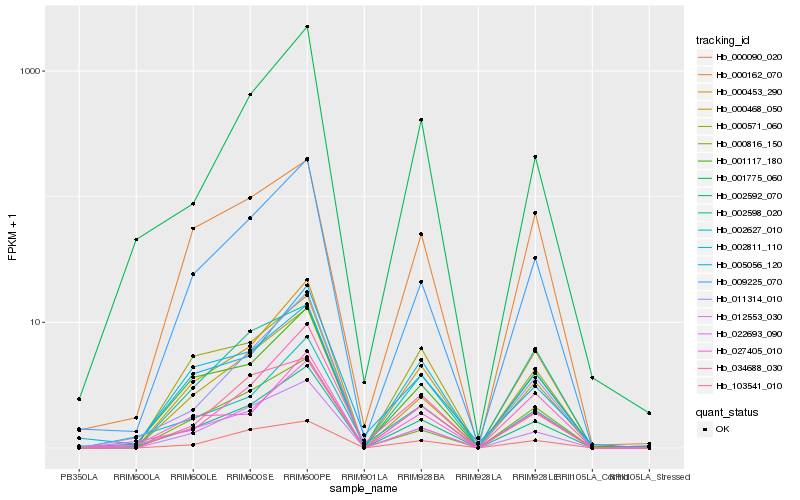
Hb_002592_070 |
0.0 |
- |
- |
hypothetical protein VITISV_020993 [Vitis vinifera] |
| 2 |
Hb_000468_050 |
0.0732585849 |
- |
- |
- |
| 3 |
Hb_000453_290 |
0.0762763415 |
- |
- |
PREDICTED: dr1-associated corepressor-like [Jatropha curcas] |
| 4 |
Hb_012553_030 |
0.0781955489 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 5 |
Hb_009225_070 |
0.0840111735 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 6 |
Hb_001117_180 |
0.0939027557 |
- |
- |
PREDICTED: uncharacterized protein LOC105629703 isoform X1 [Jatropha curcas] |
| 7 |
Hb_022693_090 |
0.0995139018 |
- |
- |
glycosyltransferase, putative [Ricinus communis] |
| 8 |
Hb_002627_010 |
0.1034191529 |
- |
- |
PREDICTED: uncharacterized protein LOC105640888 [Jatropha curcas] |
| 9 |
Hb_027405_010 |
0.107241025 |
- |
- |
PREDICTED: uncharacterized protein At3g17950-like [Populus euphratica] |
| 10 |
Hb_002811_110 |
0.1085315496 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Jatropha curcas] |
| 11 |
Hb_034688_030 |
0.1169052644 |
- |
- |
PREDICTED: uncharacterized protein LOC105642049 [Jatropha curcas] |
| 12 |
Hb_000090_020 |
0.1192059143 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 13 |
Hb_011314_010 |
0.1206654931 |
- |
- |
PREDICTED: disease resistance protein RPS6-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_103541_010 |
0.121128677 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000571_060 |
0.1229279738 |
- |
- |
PREDICTED: uncharacterized protein LOC105648112 [Jatropha curcas] |
| 16 |
Hb_000816_150 |
0.1257133359 |
- |
- |
oligopeptide transporter, putative [Ricinus communis] |
| 17 |
Hb_001775_060 |
0.1281444641 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005056_120 |
0.1297471074 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 19 |
Hb_000162_070 |
0.131882185 |
- |
- |
phospholipase C, putative [Ricinus communis] |
| 20 |
Hb_002598_020 |
0.1323744385 |
- |
- |
PREDICTED: sugar transporter ERD6-like 5 [Populus euphratica] |