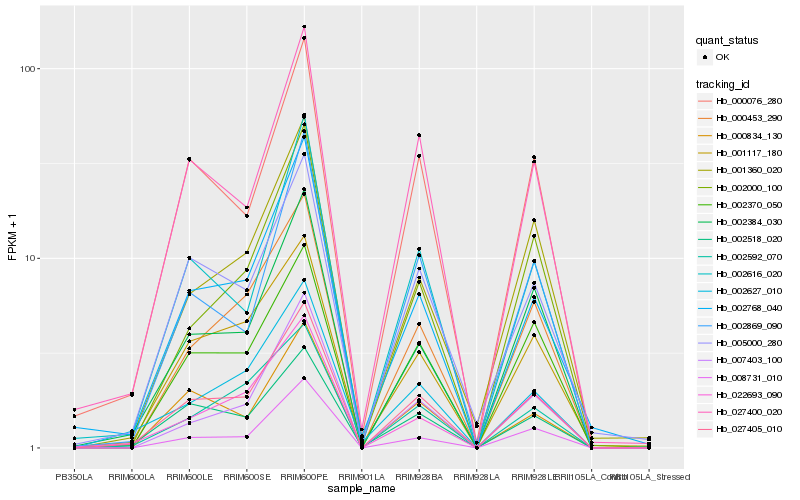
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027405_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At3g17950-like [Populus euphratica] |
| 2 |
Hb_002384_030 |
0.0885016275 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000076_280 |
0.0907756738 |
- |
- |
PREDICTED: pectinesterase-like [Populus euphratica] |
| 4 |
Hb_001117_180 |
0.0932313251 |
- |
- |
PREDICTED: uncharacterized protein LOC105629703 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000453_290 |
0.0944846837 |
- |
- |
PREDICTED: dr1-associated corepressor-like [Jatropha curcas] |
| 6 |
Hb_027400_020 |
0.0950254485 |
- |
- |
PREDICTED: uncharacterized protein At5g22580 isoform X2 [Jatropha curcas] |
| 7 |
Hb_002370_050 |
0.0971713446 |
- |
- |
hypothetical protein JCGZ_18849 [Jatropha curcas] |
| 8 |
Hb_002518_020 |
0.1011038411 |
- |
- |
PREDICTED: uncharacterized protein LOC105631164 [Jatropha curcas] |
| 9 |
Hb_022693_090 |
0.1016043001 |
- |
- |
glycosyltransferase, putative [Ricinus communis] |
| 10 |
Hb_002616_020 |
0.1025105321 |
- |
- |
PREDICTED: peroxidase 3 [Jatropha curcas] |
| 11 |
Hb_002768_040 |
0.1026069627 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_005000_280 |
0.1049932355 |
- |
- |
PREDICTED: uncharacterized protein LOC105637905 [Jatropha curcas] |
| 13 |
Hb_002627_010 |
0.1065090652 |
- |
- |
PREDICTED: uncharacterized protein LOC105640888 [Jatropha curcas] |
| 14 |
Hb_002592_070 |
0.107241025 |
- |
- |
hypothetical protein VITISV_020993 [Vitis vinifera] |
| 15 |
Hb_008731_010 |
0.1116356084 |
- |
- |
hypothetical protein POPTR_0605s00010g [Populus trichocarpa] |
| 16 |
Hb_001360_020 |
0.1130276575 |
- |
- |
PREDICTED: S-adenosylmethionine synthase 1 [Nelumbo nucifera] |
| 17 |
Hb_000834_130 |
0.1132986591 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_007403_100 |
0.1160373088 |
- |
- |
PREDICTED: probable purple acid phosphatase 20 [Jatropha curcas] |
| 19 |
Hb_002869_090 |
0.1168178441 |
- |
- |
PREDICTED: dirigent protein 22-like [Jatropha curcas] |
| 20 |
Hb_002000_100 |
0.1206308047 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming]-like [Jatropha curcas] |