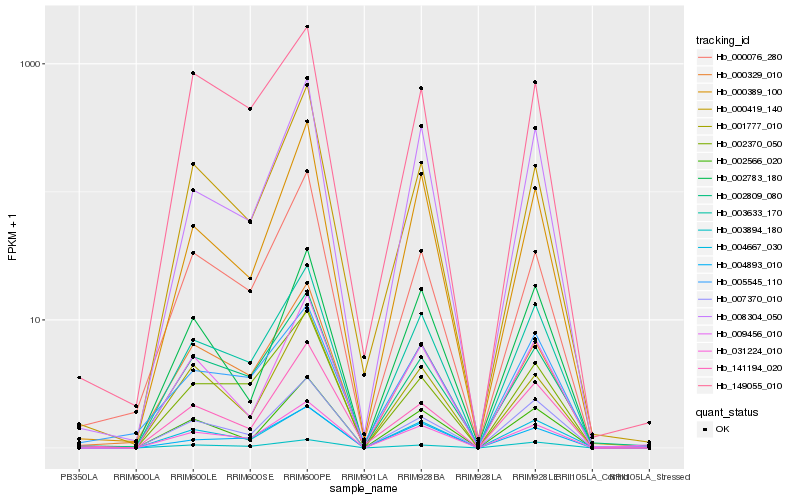
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_031224_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_003633_170 |
0.032960822 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000329_010 |
0.0797935133 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 4 |
Hb_002566_020 |
0.0799462169 |
- |
- |
hypothetical protein JCGZ_15085 [Jatropha curcas] |
| 5 |
Hb_007370_010 |
0.0812214979 |
- |
- |
PREDICTED: indole-3-pyruvate monooxygenase YUCCA6 [Jatropha curcas] |
| 6 |
Hb_002370_050 |
0.0873534252 |
- |
- |
hypothetical protein JCGZ_18849 [Jatropha curcas] |
| 7 |
Hb_002809_080 |
0.0937783066 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004893_010 |
0.0949683349 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Jatropha curcas] |
| 9 |
Hb_009456_010 |
0.0976744335 |
- |
- |
PREDICTED: cinnamoyl-CoA reductase 1-like isoform X3 [Populus euphratica] |
| 10 |
Hb_008304_050 |
0.099714105 |
- |
- |
PREDICTED: trans-cinnamate 4-monooxygenase [Jatropha curcas] |
| 11 |
Hb_004667_030 |
0.1029203042 |
- |
- |
hypothetical protein VITISV_034568 [Vitis vinifera] |
| 12 |
Hb_000389_100 |
0.1045374237 |
- |
- |
phenylalanine ammonia-lyase 1 [Manihot esculenta] |
| 13 |
Hb_003894_180 |
0.105168644 |
- |
- |
PREDICTED: uncharacterized protein LOC104245193, partial [Nicotiana sylvestris] |
| 14 |
Hb_000419_140 |
0.1065576787 |
- |
- |
Aquaporin PIP2.1, putative [Ricinus communis] |
| 15 |
Hb_149055_010 |
0.107124362 |
- |
- |
cinnamate 4-hydroxylase [Leucaena leucocephala] |
| 16 |
Hb_001777_010 |
0.1084671329 |
- |
- |
- |
| 17 |
Hb_000076_280 |
0.1116789357 |
- |
- |
PREDICTED: pectinesterase-like [Populus euphratica] |
| 18 |
Hb_141194_020 |
0.1173517227 |
- |
- |
carbohydrate transporter, putative [Ricinus communis] |
| 19 |
Hb_005545_110 |
0.1178077082 |
- |
- |
PREDICTED: protein trichome birefringence-like 33 [Jatropha curcas] |
| 20 |
Hb_002783_180 |
0.1195726401 |
- |
- |
Tubulin beta-6 chain [Gossypium arboreum] |