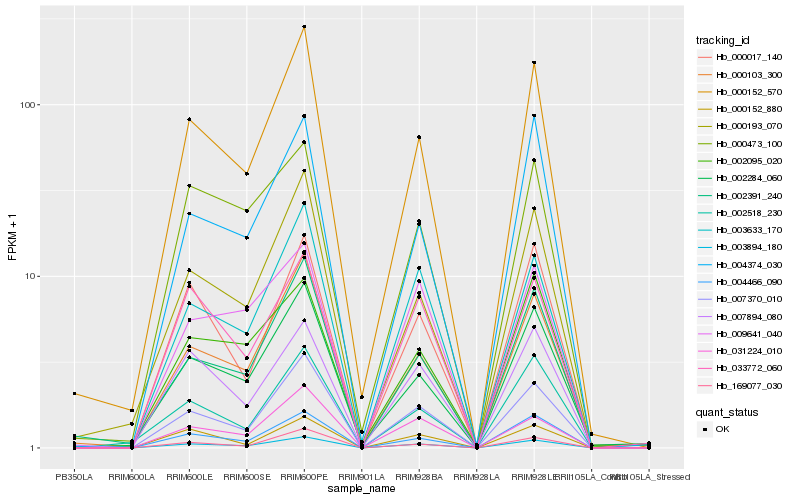
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003894_180 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104245193, partial [Nicotiana sylvestris] |
| 2 |
Hb_007370_010 |
0.0678005944 |
- |
- |
PREDICTED: indole-3-pyruvate monooxygenase YUCCA6 [Jatropha curcas] |
| 3 |
Hb_000152_570 |
0.0729014038 |
- |
- |
21 kDa protein precursor, putative [Ricinus communis] |
| 4 |
Hb_004466_090 |
0.08582707 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003633_170 |
0.0903227383 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002284_060 |
0.0924750155 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 7 |
Hb_000103_300 |
0.0948133232 |
- |
- |
PREDICTED: perakine reductase-like [Jatropha curcas] |
| 8 |
Hb_007894_080 |
0.0951209482 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000193_070 |
0.1024395691 |
- |
- |
hypothetical protein JCGZ_25373 [Jatropha curcas] |
| 10 |
Hb_000017_140 |
0.1024557136 |
- |
- |
polygalacturonase, putative [Ricinus communis] |
| 11 |
Hb_033772_060 |
0.1043117783 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_169077_030 |
0.104912303 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein RF9 [Jatropha curcas] |
| 13 |
Hb_031224_010 |
0.105168644 |
- |
- |
- |
| 14 |
Hb_002391_240 |
0.1066824931 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000152_880 |
0.1081263734 |
- |
- |
- |
| 16 |
Hb_000473_100 |
0.110733952 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 17 |
Hb_004374_030 |
0.1115296996 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 18 |
Hb_009641_040 |
0.1134469278 |
- |
- |
PREDICTED: uncharacterized protein LOC105633355 [Jatropha curcas] |
| 19 |
Hb_002095_020 |
0.114851465 |
- |
- |
PREDICTED: glutamate receptor 3.4-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_002518_230 |
0.1162413459 |
- |
- |
- |