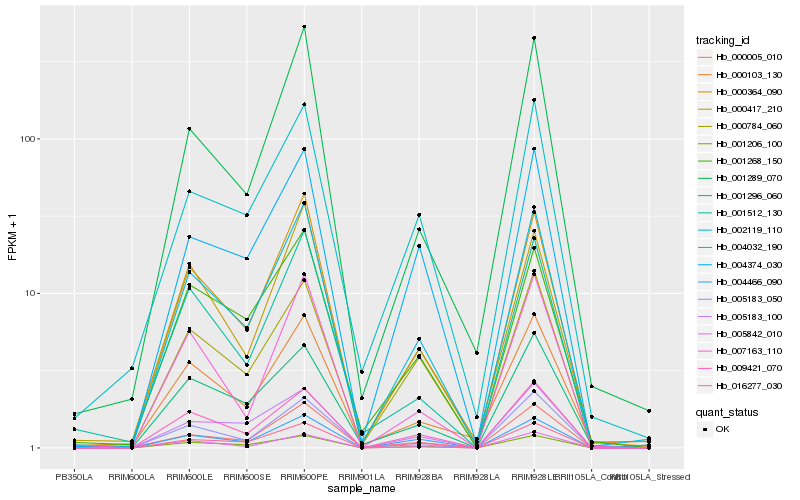
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004032_190 |
0.0 |
- |
- |
PREDICTED: probable cellulose synthase A catalytic subunit 3 [UDP-forming] isoform X1 [Jatropha curcas] |
| 2 |
Hb_000364_090 |
0.0521114161 |
- |
- |
PREDICTED: filament-like plant protein 7 isoform X2 [Jatropha curcas] |
| 3 |
Hb_016277_030 |
0.073769006 |
- |
- |
hypothetical protein JCGZ_16973 [Jatropha curcas] |
| 4 |
Hb_001206_100 |
0.0740405847 |
- |
- |
hypothetical protein PRUPE_ppa021915mg, partial [Prunus persica] |
| 5 |
Hb_009421_070 |
0.0748692726 |
- |
- |
PREDICTED: cell number regulator 1-like [Jatropha curcas] |
| 6 |
Hb_004466_090 |
0.084065282 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000784_060 |
0.0894731667 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000417_210 |
0.0929542595 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_001289_070 |
0.093346125 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 10 |
Hb_004374_030 |
0.094353379 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 11 |
Hb_005842_010 |
0.0951964411 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 12 |
Hb_001512_130 |
0.0969278915 |
- |
- |
PREDICTED: uncharacterized protein LOC105633633 [Jatropha curcas] |
| 13 |
Hb_000103_130 |
0.0993261444 |
- |
- |
PREDICTED: protein NETWORKED 1A [Jatropha curcas] |
| 14 |
Hb_000005_010 |
0.0995716285 |
- |
- |
PREDICTED: serine carboxypeptidase-like 51 [Jatropha curcas] |
| 15 |
Hb_005183_050 |
0.0998143494 |
- |
- |
hypothetical protein RCOM_1590900 [Ricinus communis] |
| 16 |
Hb_002119_110 |
0.1000583539 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 28 [Jatropha curcas] |
| 17 |
Hb_007163_110 |
0.1026097237 |
- |
- |
PREDICTED: kinesin-like protein KIF19 [Jatropha curcas] |
| 18 |
Hb_001296_060 |
0.1030482336 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 19 |
Hb_005183_100 |
0.1032355216 |
- |
- |
hypothetical protein CICLE_v10002898mg [Citrus clementina] |
| 20 |
Hb_001268_150 |
0.1036289727 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |