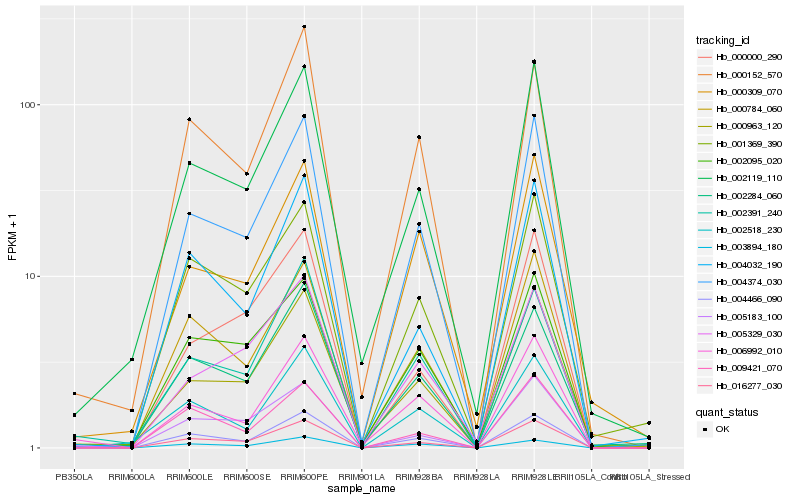
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004374_030 |
0.0 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 2 |
Hb_016277_030 |
0.045511805 |
- |
- |
hypothetical protein JCGZ_16973 [Jatropha curcas] |
| 3 |
Hb_004466_090 |
0.055198348 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000784_060 |
0.0719901275 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000963_120 |
0.0738285512 |
- |
- |
PREDICTED: uncharacterized protein LOC105628103 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002119_110 |
0.0772621979 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 28 [Jatropha curcas] |
| 7 |
Hb_002095_020 |
0.0923506307 |
- |
- |
PREDICTED: glutamate receptor 3.4-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_004032_190 |
0.094353379 |
- |
- |
PREDICTED: probable cellulose synthase A catalytic subunit 3 [UDP-forming] isoform X1 [Jatropha curcas] |
| 9 |
Hb_001369_390 |
0.0949406746 |
- |
- |
Glycerol-3-phosphate transporter, putative [Ricinus communis] |
| 10 |
Hb_005183_100 |
0.0957449241 |
- |
- |
hypothetical protein CICLE_v10002898mg [Citrus clementina] |
| 11 |
Hb_002284_060 |
0.0979915893 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 12 |
Hb_000309_070 |
0.1008759745 |
- |
- |
PREDICTED: 2-methylene-furan-3-one reductase [Cucumis melo] |
| 13 |
Hb_002518_230 |
0.1026919512 |
- |
- |
- |
| 14 |
Hb_005329_030 |
0.1062464887 |
- |
- |
PREDICTED: NAD(P)H dehydrogenase (quinone) FQR1-like [Jatropha curcas] |
| 15 |
Hb_006992_010 |
0.1072497083 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 16 |
Hb_000152_570 |
0.1076708034 |
- |
- |
21 kDa protein precursor, putative [Ricinus communis] |
| 17 |
Hb_009421_070 |
0.1082661138 |
- |
- |
PREDICTED: cell number regulator 1-like [Jatropha curcas] |
| 18 |
Hb_002391_240 |
0.108575848 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003894_180 |
0.1115296996 |
- |
- |
PREDICTED: uncharacterized protein LOC104245193, partial [Nicotiana sylvestris] |
| 20 |
Hb_000000_290 |
0.1119095969 |
- |
- |
PREDICTED: probable 1-acyl-sn-glycerol-3-phosphate acyltransferase 4 [Jatropha curcas] |