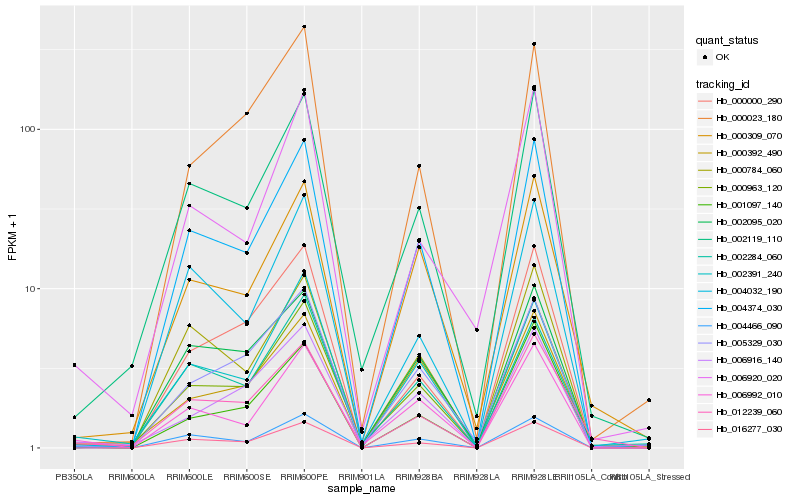
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000963_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628103 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002119_110 |
0.053785768 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 28 [Jatropha curcas] |
| 3 |
Hb_004374_030 |
0.0738285512 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 4 |
Hb_016277_030 |
0.0917914346 |
- |
- |
hypothetical protein JCGZ_16973 [Jatropha curcas] |
| 5 |
Hb_006916_140 |
0.0953016841 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like [Jatropha curcas] |
| 6 |
Hb_000309_070 |
0.0961524885 |
- |
- |
PREDICTED: 2-methylene-furan-3-one reductase [Cucumis melo] |
| 7 |
Hb_012239_060 |
0.0976203179 |
- |
- |
PREDICTED: nudix hydrolase 18, mitochondrial [Jatropha curcas] |
| 8 |
Hb_006992_010 |
0.1031893141 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 9 |
Hb_000000_290 |
0.1055669143 |
- |
- |
PREDICTED: probable 1-acyl-sn-glycerol-3-phosphate acyltransferase 4 [Jatropha curcas] |
| 10 |
Hb_001097_140 |
0.1094776055 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002284_060 |
0.1100203589 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 12 |
Hb_005329_030 |
0.1104063308 |
- |
- |
PREDICTED: NAD(P)H dehydrogenase (quinone) FQR1-like [Jatropha curcas] |
| 13 |
Hb_004466_090 |
0.1117639074 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000784_060 |
0.1132569787 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000023_180 |
0.1137243363 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 16 |
Hb_000392_490 |
0.1151970506 |
- |
- |
PREDICTED: uncharacterized protein LOC105640495 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002095_020 |
0.1152960196 |
- |
- |
PREDICTED: glutamate receptor 3.4-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_002391_240 |
0.1171024885 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004032_190 |
0.1178129231 |
- |
- |
PREDICTED: probable cellulose synthase A catalytic subunit 3 [UDP-forming] isoform X1 [Jatropha curcas] |
| 20 |
Hb_006920_020 |
0.1181877665 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |