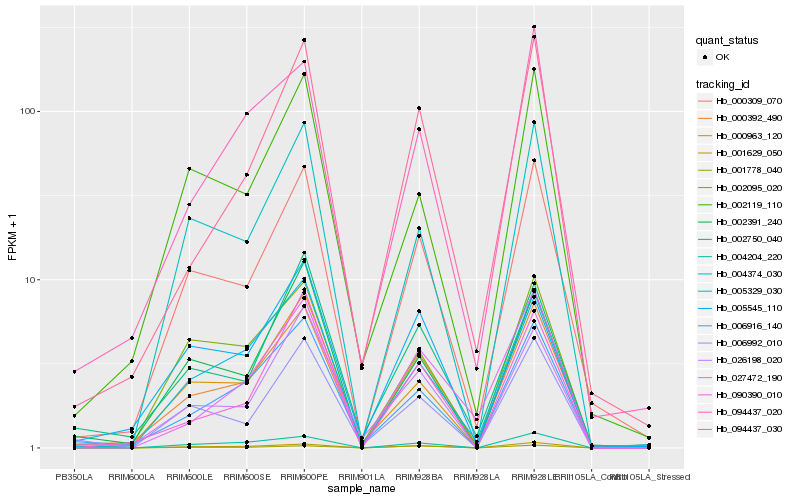
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000392_490 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640495 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000309_070 |
0.074402043 |
- |
- |
PREDICTED: 2-methylene-furan-3-one reductase [Cucumis melo] |
| 3 |
Hb_006916_140 |
0.1084715978 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like [Jatropha curcas] |
| 4 |
Hb_001629_050 |
0.1097427374 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 5 |
Hb_005329_030 |
0.1113576168 |
- |
- |
PREDICTED: NAD(P)H dehydrogenase (quinone) FQR1-like [Jatropha curcas] |
| 6 |
Hb_026198_020 |
0.1146846711 |
- |
- |
PREDICTED: uncharacterized protein LOC105634368 [Jatropha curcas] |
| 7 |
Hb_000963_120 |
0.1151970506 |
- |
- |
PREDICTED: uncharacterized protein LOC105628103 isoform X1 [Jatropha curcas] |
| 8 |
Hb_094437_030 |
0.1183608781 |
- |
- |
hypothetical protein AMTR_s00420p00013340 [Amborella trichopoda] |
| 9 |
Hb_094437_020 |
0.1210485533 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 10 |
Hb_004204_220 |
0.1224576798 |
- |
- |
- |
| 11 |
Hb_002750_040 |
0.1230466142 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g67200 [Jatropha curcas] |
| 12 |
Hb_004374_030 |
0.1284693215 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 13 |
Hb_002095_020 |
0.1296767884 |
- |
- |
PREDICTED: glutamate receptor 3.4-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_006992_010 |
0.1300888891 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 15 |
Hb_002119_110 |
0.1310501207 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 28 [Jatropha curcas] |
| 16 |
Hb_005545_110 |
0.139820279 |
- |
- |
PREDICTED: protein trichome birefringence-like 33 [Jatropha curcas] |
| 17 |
Hb_090390_010 |
0.1410826936 |
- |
- |
hypothetical protein POPTR_0006s28850g [Populus trichocarpa] |
| 18 |
Hb_002391_240 |
0.1414148612 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_027472_190 |
0.1434460714 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 20 |
Hb_001778_040 |
0.1447016757 |
- |
- |
hypothetical protein VITISV_021035 [Vitis vinifera] |