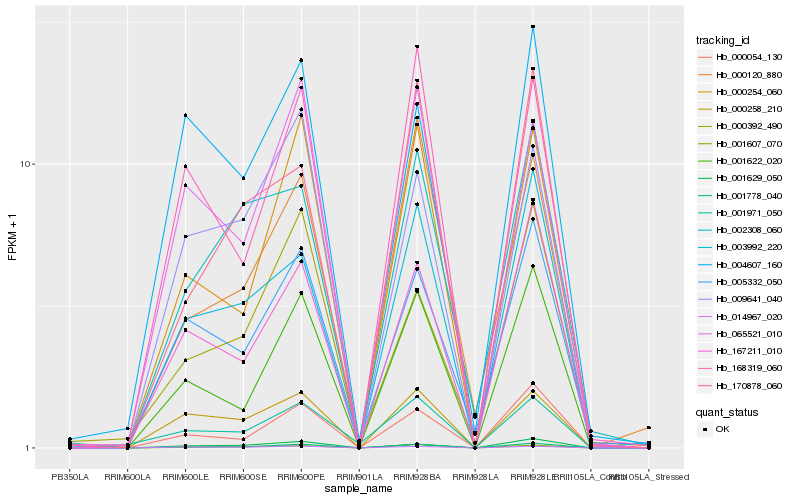
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001778_040 |
0.0 |
- |
- |
hypothetical protein VITISV_021035 [Vitis vinifera] |
| 2 |
Hb_001622_020 |
0.0690428657 |
- |
- |
- |
| 3 |
Hb_000054_130 |
0.0806819903 |
- |
- |
hypothetical protein JCGZ_18787 [Jatropha curcas] |
| 4 |
Hb_167211_010 |
0.0883978942 |
- |
- |
PREDICTED: glutamate receptor 3.4-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_000254_060 |
0.0896683029 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003992_220 |
0.1083673691 |
- |
- |
s-receptor kinase, putative [Ricinus communis] |
| 7 |
Hb_001629_050 |
0.1148583051 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 8 |
Hb_005332_050 |
0.1179593226 |
- |
- |
hypothetical protein POPTR_0017s10650g [Populus trichocarpa] |
| 9 |
Hb_000258_210 |
0.1226100536 |
- |
- |
prenyl-dependent CAAX protease, putative [Ricinus communis] |
| 10 |
Hb_001607_070 |
0.1266810097 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 11 |
Hb_168319_060 |
0.127230005 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Populus euphratica] |
| 12 |
Hb_014967_020 |
0.1305722091 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 13 |
Hb_065521_010 |
0.132980692 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL5 [Jatropha curcas] |
| 14 |
Hb_009641_040 |
0.1383976555 |
- |
- |
PREDICTED: uncharacterized protein LOC105633355 [Jatropha curcas] |
| 15 |
Hb_170878_060 |
0.1393091193 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 16 |
Hb_001971_050 |
0.140931734 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 17 |
Hb_000120_880 |
0.1430348865 |
- |
- |
PREDICTED: disease resistance response protein 206-like [Jatropha curcas] |
| 18 |
Hb_000392_490 |
0.1447016757 |
- |
- |
PREDICTED: uncharacterized protein LOC105640495 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004607_160 |
0.1470999497 |
- |
- |
PREDICTED: uncharacterized protein LOC105134931 [Populus euphratica] |
| 20 |
Hb_002308_060 |
0.1490156389 |
- |
- |
PREDICTED: cytochrome P450 704C1-like [Jatropha curcas] |