| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
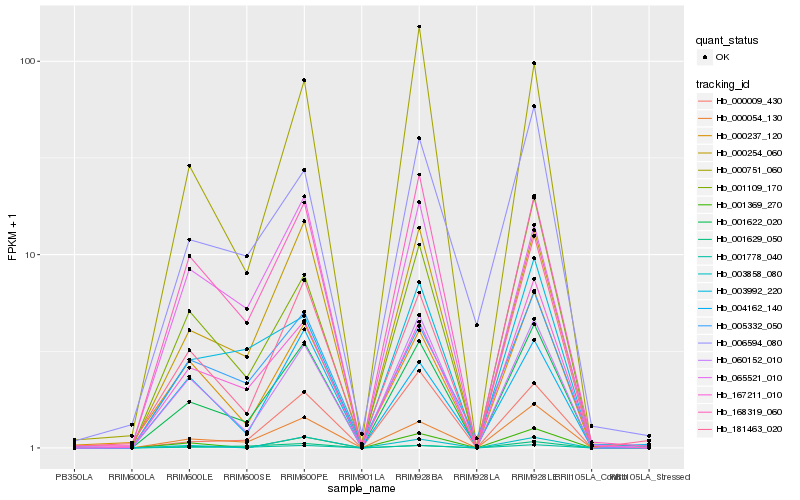
Hb_001622_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_000054_130 |
0.059456166 |
- |
- |
hypothetical protein JCGZ_18787 [Jatropha curcas] |
| 3 |
Hb_001778_040 |
0.0690428657 |
- |
- |
hypothetical protein VITISV_021035 [Vitis vinifera] |
| 4 |
Hb_000254_060 |
0.0783087449 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_167211_010 |
0.0953744656 |
- |
- |
PREDICTED: glutamate receptor 3.4-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_060152_010 |
0.1029263605 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 7 |
Hb_168319_060 |
0.1147785888 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Populus euphratica] |
| 8 |
Hb_181463_020 |
0.1155781892 |
- |
- |
hypothetical protein JCGZ_26765 [Jatropha curcas] |
| 9 |
Hb_000237_120 |
0.1183779817 |
- |
- |
PREDICTED: beta-galactosidase 3-like [Jatropha curcas] |
| 10 |
Hb_001109_170 |
0.1199618959 |
- |
- |
PREDICTED: urea-proton symporter DUR3 isoform X2 [Jatropha curcas] |
| 11 |
Hb_005332_050 |
0.1343147736 |
- |
- |
hypothetical protein POPTR_0017s10650g [Populus trichocarpa] |
| 12 |
Hb_000751_060 |
0.1407900241 |
- |
- |
PREDICTED: extensin-2-like, partial [Sesamum indicum] |
| 13 |
Hb_003992_220 |
0.14156278 |
- |
- |
s-receptor kinase, putative [Ricinus communis] |
| 14 |
Hb_065521_010 |
0.1464957877 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL5 [Jatropha curcas] |
| 15 |
Hb_004162_140 |
0.1505378339 |
- |
- |
PREDICTED: serine carboxypeptidase-like 51 [Jatropha curcas] |
| 16 |
Hb_001369_270 |
0.1507168307 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_003858_080 |
0.1507352988 |
- |
- |
Germin-like protein 2 [Theobroma cacao] |
| 18 |
Hb_006594_080 |
0.1524790213 |
- |
- |
beta-glucosidase, putative [Ricinus communis] |
| 19 |
Hb_000009_430 |
0.1553763249 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001629_050 |
0.1570887438 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |