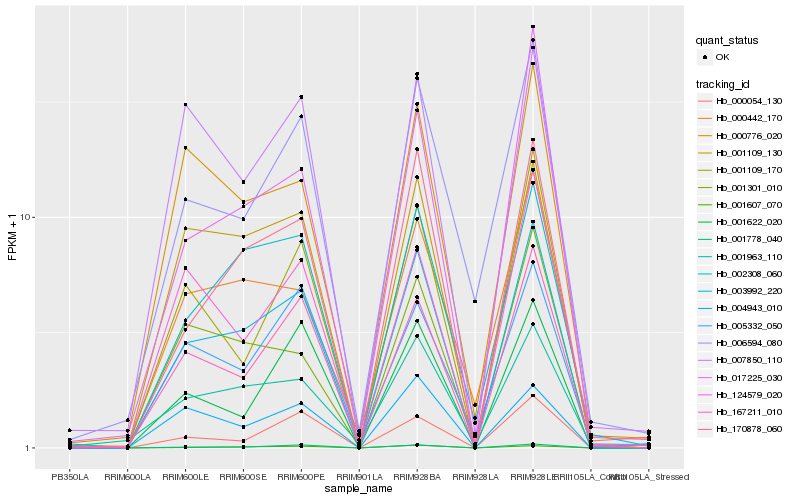
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003992_220 |
0.0 |
- |
- |
s-receptor kinase, putative [Ricinus communis] |
| 2 |
Hb_170878_060 |
0.0955218867 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 3 |
Hb_167211_010 |
0.1051358321 |
- |
- |
PREDICTED: glutamate receptor 3.4-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_001778_040 |
0.1083673691 |
- |
- |
hypothetical protein VITISV_021035 [Vitis vinifera] |
| 5 |
Hb_002308_060 |
0.1135672777 |
- |
- |
PREDICTED: cytochrome P450 704C1-like [Jatropha curcas] |
| 6 |
Hb_001109_130 |
0.1152153402 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 136 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001963_110 |
0.1226013081 |
- |
- |
- |
| 8 |
Hb_000776_020 |
0.1281863861 |
transcription factor |
TF Family: M-type |
hypothetical protein JCGZ_25594 [Jatropha curcas] |
| 9 |
Hb_001607_070 |
0.1285373665 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 10 |
Hb_017225_030 |
0.130773995 |
- |
- |
PREDICTED: probable cytosolic oligopeptidase A [Jatropha curcas] |
| 11 |
Hb_006594_080 |
0.131626267 |
- |
- |
beta-glucosidase, putative [Ricinus communis] |
| 12 |
Hb_000442_170 |
0.1320809586 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 13 |
Hb_001109_170 |
0.1346189444 |
- |
- |
PREDICTED: urea-proton symporter DUR3 isoform X2 [Jatropha curcas] |
| 14 |
Hb_007850_110 |
0.1368572635 |
- |
- |
MADS-box transcription factor 3 [Hevea brasiliensis] |
| 15 |
Hb_001301_010 |
0.1384875632 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 16 |
Hb_004943_010 |
0.1408040311 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 17 |
Hb_001622_020 |
0.14156278 |
- |
- |
- |
| 18 |
Hb_000054_130 |
0.1426264846 |
- |
- |
hypothetical protein JCGZ_18787 [Jatropha curcas] |
| 19 |
Hb_005332_050 |
0.14280854 |
- |
- |
hypothetical protein POPTR_0017s10650g [Populus trichocarpa] |
| 20 |
Hb_124579_020 |
0.1435077216 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Jatropha curcas] |