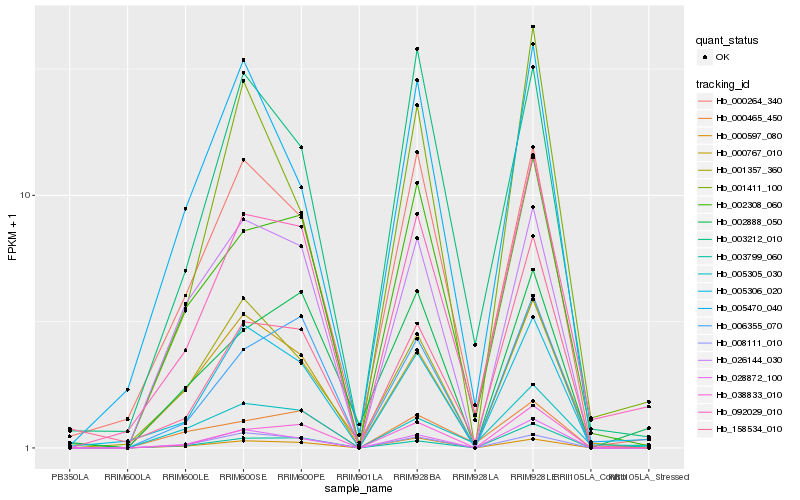
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_092029_010 |
0.0 |
- |
- |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 2 |
Hb_005305_030 |
0.1201998275 |
- |
- |
hypothetical protein JCGZ_25922 [Jatropha curcas] |
| 3 |
Hb_002308_060 |
0.1266283172 |
- |
- |
PREDICTED: cytochrome P450 704C1-like [Jatropha curcas] |
| 4 |
Hb_000264_340 |
0.1441469035 |
- |
- |
PREDICTED: callose synthase 11-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_038833_010 |
0.1497500146 |
- |
- |
polyprotein [Ananas comosus] |
| 6 |
Hb_000465_450 |
0.1500655917 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 7 |
Hb_005306_020 |
0.1520544527 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86 [Jatropha curcas] |
| 8 |
Hb_028872_100 |
0.1560131444 |
- |
- |
PREDICTED: uncharacterized protein LOC102707106 [Oryza brachyantha] |
| 9 |
Hb_158534_010 |
0.1579196356 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 10 |
Hb_003799_060 |
0.15874483 |
- |
- |
hypothetical protein POPTR_0005s22800g [Populus trichocarpa] |
| 11 |
Hb_000767_010 |
0.1597010548 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like isoform X2 [Solanum lycopersicum] |
| 12 |
Hb_006355_070 |
0.1632794497 |
transcription factor |
TF Family: bHLH |
Phytochrome-interacting factor, putative [Ricinus communis] |
| 13 |
Hb_003212_010 |
0.1668230051 |
- |
- |
sucrose transporter 4 [Hevea brasiliensis] |
| 14 |
Hb_001357_360 |
0.1724786998 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 15 |
Hb_005470_040 |
0.1740912095 |
- |
- |
hypothetical protein POPTR_0015s09760g [Populus trichocarpa] |
| 16 |
Hb_000597_080 |
0.1753811986 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_008111_010 |
0.1765354174 |
- |
- |
hypothetical protein VITISV_021527 [Vitis vinifera] |
| 18 |
Hb_026144_030 |
0.1766304208 |
- |
- |
PREDICTED: high affinity nitrate transporter 2.5 [Jatropha curcas] |
| 19 |
Hb_001411_100 |
0.1770053746 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein 2 precursor, putative [Ricinus communis] |
| 20 |
Hb_002888_050 |
0.177758478 |
- |
- |
PREDICTED: phospholipase SGR2 isoform X1 [Jatropha curcas] |