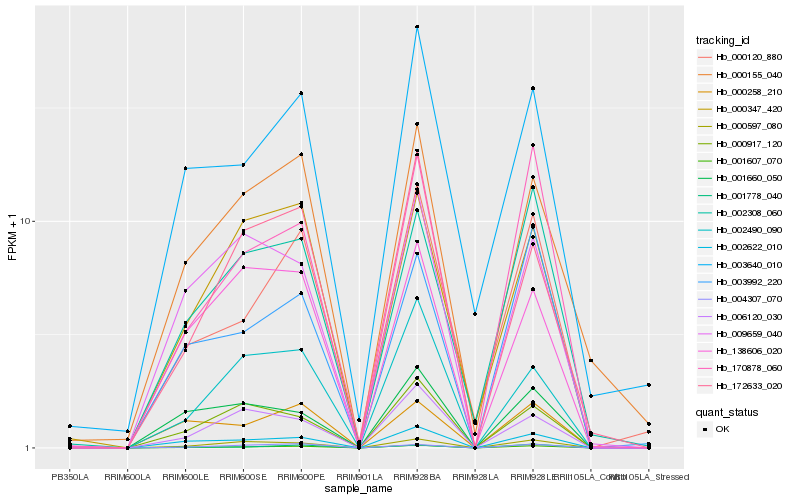
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001607_070 |
0.0 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 2 |
Hb_002622_010 |
0.0912729193 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 3 |
Hb_170878_060 |
0.0997511108 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 4 |
Hb_000597_080 |
0.1042588759 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_000120_880 |
0.1153693945 |
- |
- |
PREDICTED: disease resistance response protein 206-like [Jatropha curcas] |
| 6 |
Hb_002308_060 |
0.1251054427 |
- |
- |
PREDICTED: cytochrome P450 704C1-like [Jatropha curcas] |
| 7 |
Hb_001778_040 |
0.1266810097 |
- |
- |
hypothetical protein VITISV_021035 [Vitis vinifera] |
| 8 |
Hb_003992_220 |
0.1285373665 |
- |
- |
s-receptor kinase, putative [Ricinus communis] |
| 9 |
Hb_004307_070 |
0.1309598767 |
- |
- |
kinase, putative [Ricinus communis] |
| 10 |
Hb_000917_120 |
0.1353197808 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000258_210 |
0.1371539609 |
- |
- |
prenyl-dependent CAAX protease, putative [Ricinus communis] |
| 12 |
Hb_002490_090 |
0.1385405428 |
- |
- |
PREDICTED: uncharacterized protein LOC105133003 [Populus euphratica] |
| 13 |
Hb_000347_420 |
0.139892549 |
- |
- |
hypothetical protein JCGZ_24696 [Jatropha curcas] |
| 14 |
Hb_006120_030 |
0.1432212238 |
- |
- |
- |
| 15 |
Hb_001660_050 |
0.144303473 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Eucalyptus grandis] |
| 16 |
Hb_000155_040 |
0.144371896 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 17 |
Hb_172633_020 |
0.146156267 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g56140 [Populus euphratica] |
| 18 |
Hb_009659_040 |
0.1463128354 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein NUTCRACKER [Populus euphratica] |
| 19 |
Hb_138606_020 |
0.1496553368 |
- |
- |
Nitrate transporter2.5 [Theobroma cacao] |
| 20 |
Hb_003640_010 |
0.1522084709 |
- |
- |
interferon-induced GTP-binding protein mx, putative [Ricinus communis] |