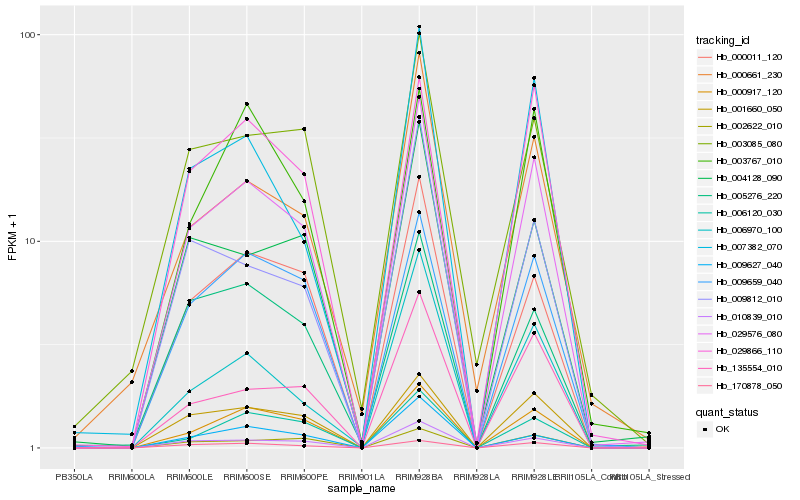
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029576_080 |
0.0 |
- |
- |
Aspartic proteinase precursor, putative [Ricinus communis] |
| 2 |
Hb_001660_050 |
0.0784999329 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Eucalyptus grandis] |
| 3 |
Hb_010839_010 |
0.0838758461 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 4 |
Hb_000917_120 |
0.0908821793 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_009659_040 |
0.1044359969 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein NUTCRACKER [Populus euphratica] |
| 6 |
Hb_000011_120 |
0.1045369251 |
- |
- |
PREDICTED: ATPase 10, plasma membrane-type-like [Populus euphratica] |
| 7 |
Hb_170878_050 |
0.1053645762 |
- |
- |
retrotransposon protein, putative, unclassified [Oryza sativa Japonica Group] |
| 8 |
Hb_029866_110 |
0.1104159242 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter C family member 10-like [Jatropha curcas] |
| 9 |
Hb_135554_010 |
0.1107625032 |
- |
- |
Insulin-degrading enzyme, putative [Ricinus communis] |
| 10 |
Hb_002622_010 |
0.1127432155 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 11 |
Hb_007382_070 |
0.1137211666 |
- |
- |
PREDICTED: MAPK-interacting and spindle-stabilizing protein-like [Jatropha curcas] |
| 12 |
Hb_006120_030 |
0.1197622433 |
- |
- |
- |
| 13 |
Hb_004128_090 |
0.1250606435 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 44 [Jatropha curcas] |
| 14 |
Hb_005276_220 |
0.1271617185 |
transcription factor |
TF Family: bHLH |
PREDICTED: basic helix-loop-helix protein A [Jatropha curcas] |
| 15 |
Hb_000661_230 |
0.1278781358 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 22-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_003085_080 |
0.1286709488 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 17 |
Hb_006970_100 |
0.132421125 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 18 |
Hb_009812_010 |
0.1347317412 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 19 |
Hb_009627_040 |
0.1362753129 |
- |
- |
Lipase class 3-related protein, putative [Theobroma cacao] |
| 20 |
Hb_003767_010 |
0.1370657712 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 [Jatropha curcas] |