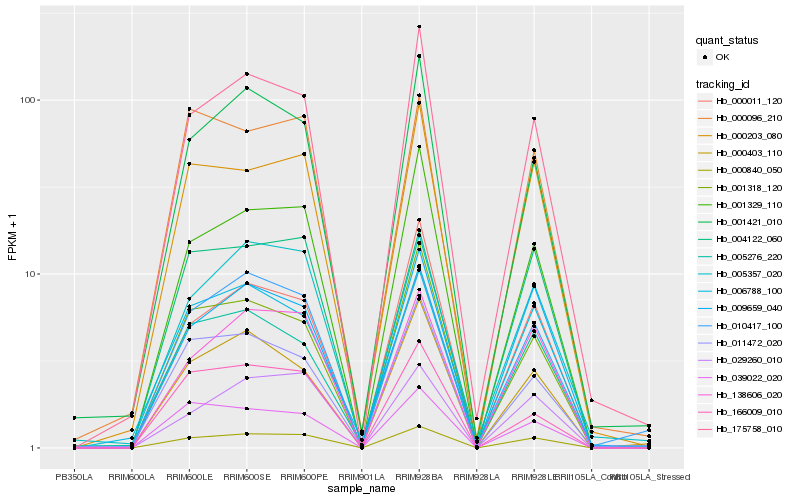
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000840_050 |
0.0 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Vitis vinifera] |
| 2 |
Hb_138606_020 |
0.0731322663 |
- |
- |
Nitrate transporter2.5 [Theobroma cacao] |
| 3 |
Hb_175758_010 |
0.08084866 |
- |
- |
PREDICTED: aspartic proteinase A1-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_000203_080 |
0.081429256 |
- |
- |
Xylem bark cysteine peptidase 3 isoform 1 [Theobroma cacao] |
| 5 |
Hb_001329_110 |
0.0857518778 |
- |
- |
PREDICTED: uncharacterized protein LOC105642675 [Jatropha curcas] |
| 6 |
Hb_009659_040 |
0.0944048937 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein NUTCRACKER [Populus euphratica] |
| 7 |
Hb_010417_100 |
0.0953196019 |
- |
- |
PREDICTED: respiratory burst oxidase homolog protein A [Jatropha curcas] |
| 8 |
Hb_029260_010 |
0.0957687969 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 9 |
Hb_004122_060 |
0.0959251552 |
- |
- |
PREDICTED: uncharacterized acetyltransferase At3g50280-like [Jatropha curcas] |
| 10 |
Hb_039022_020 |
0.0963756962 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 11 |
Hb_001421_010 |
0.0985576445 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator-like APRR2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_006788_100 |
0.101072042 |
- |
- |
PREDICTED: uncharacterized protein LOC105649919 isoform X1 [Jatropha curcas] |
| 13 |
Hb_005357_020 |
0.1034811685 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001318_120 |
0.1080833037 |
- |
- |
PREDICTED: uncharacterized protein LOC105642675 [Jatropha curcas] |
| 15 |
Hb_005276_220 |
0.1110227077 |
transcription factor |
TF Family: bHLH |
PREDICTED: basic helix-loop-helix protein A [Jatropha curcas] |
| 16 |
Hb_000096_210 |
0.1119745147 |
- |
- |
PREDICTED: probable carboxylesterase 7 [Jatropha curcas] |
| 17 |
Hb_000011_120 |
0.1169832759 |
- |
- |
PREDICTED: ATPase 10, plasma membrane-type-like [Populus euphratica] |
| 18 |
Hb_000403_110 |
0.1179820436 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 51 [Jatropha curcas] |
| 19 |
Hb_011472_020 |
0.1180733062 |
- |
- |
PREDICTED: mucin-17-like [Jatropha curcas] |
| 20 |
Hb_166009_010 |
0.118435211 |
- |
- |
carbonyl reductase, putative [Ricinus communis] |